In the rapidly evolving world of plant genomics, a new review published in the journal *Current Plant Biology* (translated from Persian as *Current Plant Biology*) is set to transform how researchers integrate and interpret RNA sequencing (RNA-Seq) data. Led by Bahman Panahi from the Department of Genomics at the Agricultural Biotechnology Research Institute of Iran (ABRII), this comprehensive review highlights the challenges and solutions in unifying RNA-Seq data from diverse studies, offering a roadmap for more consistent and reliable transcriptomic analyses.
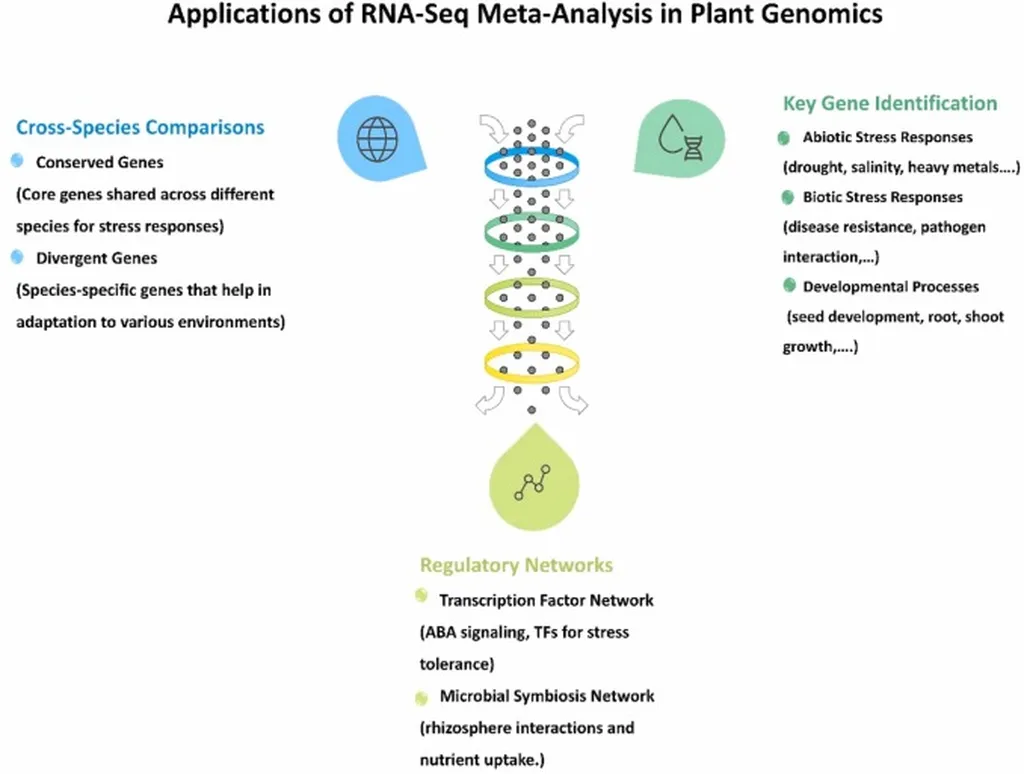
RNA-Seq has revolutionized plant genomics by providing detailed insights into gene expression across various conditions. However, the variability in experimental designs, sequencing platforms, and data processing workflows has made it difficult to compare and integrate data from different studies. This inconsistency limits the broader applicability of transcriptomic datasets, hindering progress in areas such as precision breeding and stress-response studies.
Panahi and his team delve into the methodologies that address these challenges, emphasizing the importance of data normalization, statistical frameworks for aggregating results, and computational tools that reduce inter-study variability. “By standardizing preprocessing strategies, such as batch effect correction and gene annotation pipelines, we can facilitate more reliable cross-study comparisons,” Panahi explains. These advancements not only enhance the identification of differentially expressed genes (DEGs) but also improve functional annotation and uncover conserved regulatory mechanisms across plant species.
The practical significance of RNA-Seq meta-analysis extends to various agricultural applications. For instance, in precision breeding programs, the ability to identify consistent DEGs across multiple studies can lead to more targeted and effective genetic improvements. Similarly, in stress-response studies, understanding conserved regulatory mechanisms can help develop crops that are more resilient to environmental challenges.
Panahi’s review also outlines key considerations and recommended practices for researchers implementing meta-analysis, providing a valuable resource for those looking to leverage these techniques in their work. “As transcriptomic datasets continue to expand, meta-analysis will play a crucial role in advancing our understanding of plant biology and its application in agriculture,” Panahi notes.
The review concludes by highlighting the need for standardized protocols and promoting multi-omics integration to unlock deeper insights. By addressing these challenges, researchers can pave the way for more robust and comprehensive analyses, ultimately driving innovation in plant genomics and agriculture.
This research not only shapes the future of plant genomics but also has significant implications for the energy sector. As the demand for sustainable and renewable energy sources grows, the development of bioenergy crops that are more efficient and resilient becomes increasingly important. By leveraging RNA-Seq meta-analysis, researchers can identify genetic traits that enhance biomass production and stress tolerance, contributing to the development of more sustainable bioenergy solutions.
In summary, Panahi’s review offers a compelling vision for the future of plant genomics, emphasizing the importance of integrating and standardizing RNA-Seq data. As researchers adopt these methodologies, they will unlock new insights into plant biology, driving advancements in agriculture and the energy sector.