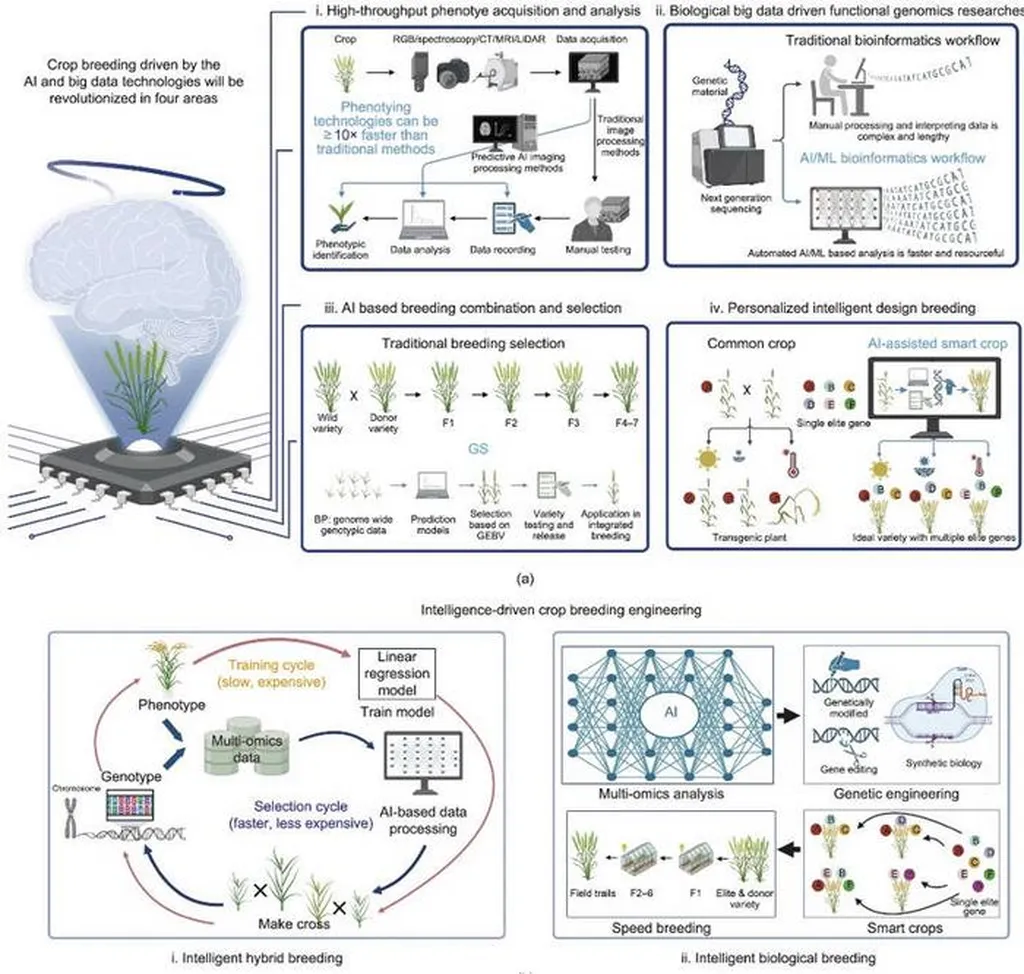
In the ever-evolving landscape of agricultural technology, a groundbreaking development has emerged from the labs of Zhejiang University. Researchers, led by Shuchang Zhou of the Zhejiang Provincial Key Laboratory of Crop Genetic Resources, have introduced CropARNet, a novel deep learning framework designed to revolutionize genomic selection (GS) in crop breeding. This innovation, detailed in a recent study published in the journal *Crop Design* (translated from Chinese as “Crop Design”), promises to enhance the efficiency and accuracy of predicting complex traits in crops, potentially reshaping the future of molecular breeding.
Genomic selection has long been a cornerstone of modern crop breeding, utilizing genome-wide markers to predict and improve complex traits. However, the integration of deep learning techniques has recently shown promise in further enhancing prediction accuracy. CropARNet stands out by combining a self-attention mechanism with a deep residual network, offering a robust and powerful tool for breeders.
“CropARNet represents a significant advancement in our ability to predict complex traits in crops,” said Shuchang Zhou, lead author of the study. “By leveraging deep learning, we can achieve higher prediction accuracy, which is crucial for accelerating the breeding process and developing crops with desirable traits.”
The study systematically evaluated CropARNet’s performance across 53 key agronomic traits in four major crops: rice, maize, cotton, and millet. When compared to established models such as GBLUP, DNNGP, XGBoost, and CropFormer, CropARNet ranked first in prediction accuracy for 29 of the 53 traits and consistently placed among the top performers for the remaining traits. This remarkable performance underscores the potential of CropARNet to become a standard tool in the field of molecular breeding.
One of the most compelling aspects of CropARNet is its ability to predict phenotypes using transcriptomic data. This capability opens new avenues for understanding and manipulating gene expression, ultimately leading to more precise and efficient breeding programs.
The commercial implications of this research are substantial. As the global demand for food continues to rise, the need for high-yield, resilient crops becomes increasingly critical. CropARNet’s enhanced prediction accuracy can significantly accelerate the development of new crop varieties, reducing the time and resources required for traditional breeding methods. This efficiency gain can translate into substantial cost savings and increased productivity for the agricultural sector.
Moreover, the integration of deep learning in genomic selection aligns with the broader trend of digital transformation in agriculture. As precision farming and data-driven decision-making become more prevalent, tools like CropARNet will play a pivotal role in optimizing crop breeding programs.
The availability of CropARNet software and illustrative examples on GitHub further democratizes access to this cutting-edge technology, enabling researchers and breeders worldwide to leverage its capabilities. This open-access approach fosters collaboration and innovation, accelerating the adoption of deep learning techniques in agricultural research.
In summary, CropARNet represents a significant leap forward in the field of molecular breeding. Its robust performance, combined with the potential for widespread adoption, positions it as a key player in the future of crop improvement. As the agricultural sector continues to evolve, innovations like CropARNet will be instrumental in meeting the challenges of food security and sustainability.
For those interested in exploring the full potential of CropARNet, the software and additional resources are available at the provided GitHub link. This research, published in *Crop Design*, marks a pivotal moment in the intersection of deep learning and agricultural science, paving the way for a more efficient and productive future in crop breeding.