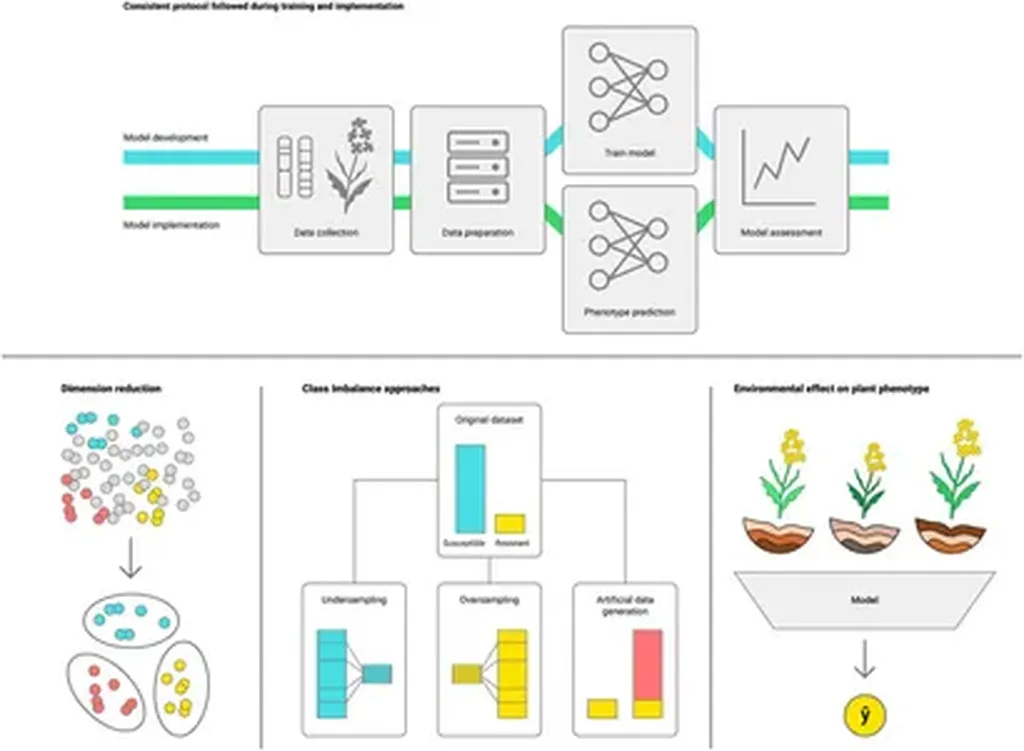
In the ever-evolving landscape of modern agriculture, precision and predictability are key to maximizing crop yields and ensuring food security. A recent study published in *Scientific Reports* offers a promising advancement in this arena, demonstrating how the integration of environmental data and machine learning can significantly enhance the accuracy of genotype predictions in multi-environment trials. This breakthrough could have profound implications for plant breeders and farmers alike, particularly in regions with diverse environmental conditions.
The research, led by Gabriel M. Blasques from the Department of Agronomy at the Federal University of Viçosa, focuses on refining the Geographic Information Systems-Factor Analytic (GIS-FA) method. By incorporating Random Forest Spatial Interpolation for improved environmental data interpolation and optimizing spatial sampling to exclude non-agricultural areas, the team achieved a notable 15.2% increase in prediction accuracy. This enhancement allows for more reliable genotype performance predictions, a critical factor in the selection and recommendation of crop varieties.
“Our goal was to make the GIS-FA method more robust and accurate,” Blasques explained. “By leveraging machine learning-based interpolation and spatial optimization, we can now provide breeders with a more comprehensive understanding of how different genotypes will perform across various environments.”
The study applied the improved GIS-FA framework to common bean trials conducted across 23 environments in São Paulo, Brazil, evaluating 59 genotypes from the “Carioca” and “Black” market classes. The results were striking: the empirical Best Linear Unbiased Predictions (eBLUPs) accuracy improved from 0.46 to 0.53 in leave-one-out cross-validation. This leap in accuracy is not just a statistical victory; it translates to real-world benefits for farmers and breeders.
One of the most exciting aspects of this research is its potential to revolutionize variety recommendation. High-resolution thematic maps generated through GIS-FA can now provide a detailed spatial view of genotype performance across the entire Target Population of Environments (TPE). This means that breeders can make more informed decisions about which varieties to recommend for specific regions, ultimately leading to higher yields and better resource utilization.
“This method allows us to predict genotype performance in untested environments with greater confidence,” Blasques noted. “It’s a game-changer for breeders who are constantly seeking to optimize their selection strategies.”
The commercial impact of this research is substantial. For farmers, the ability to select the most suitable genotypes for their specific environmental conditions can lead to increased productivity and reduced risk. For breeders, the enhanced predictive power of the GIS-FA method can streamline the selection process, saving time and resources. Moreover, the integration of GIS-FA with Factor Analytic Selection Tools provides a more nuanced understanding of stability and adaptability metrics, further refining the recommendation process.
Looking ahead, the success of this study opens the door to further advancements in the field of enviromics. As machine learning and spatial optimization techniques continue to evolve, their integration into agricultural research is likely to become even more sophisticated. This could lead to the development of even more accurate and reliable predictive models, ultimately benefiting the entire agriculture sector.
In conclusion, the research led by Blasques and his team represents a significant step forward in the quest for precision agriculture. By enhancing the GIS-FA method with cutting-edge machine learning techniques, they have provided a powerful tool for breeders and farmers to navigate the complexities of genotype-by-environment interactions. As the agriculture sector continues to face the challenges of climate change and increasing demand for food, such innovations will be crucial in ensuring sustainable and productive farming practices.