In the intricate world of gene regulation and disease mechanisms, a new tool has emerged that could significantly enhance our understanding of SUMOylation, a process critical to various biological functions. Researchers have developed SUMO-LMNet, a deep learning-based framework designed to precisely predict SUMO1 and SUMO2 modification sites, addressing a longstanding challenge in the field.
SUMOylation, the process by which Small Ubiquitin-like Modifiers (SUMOs) attach to target proteins, plays a pivotal role in regulating protein function, localization, and interactions. However, distinguishing between SUMO1 and SUMO2 modifications has been a major hurdle due to their structural similarities. Conventional prediction models often struggle with this differentiation, limiting their applicability in biological research.
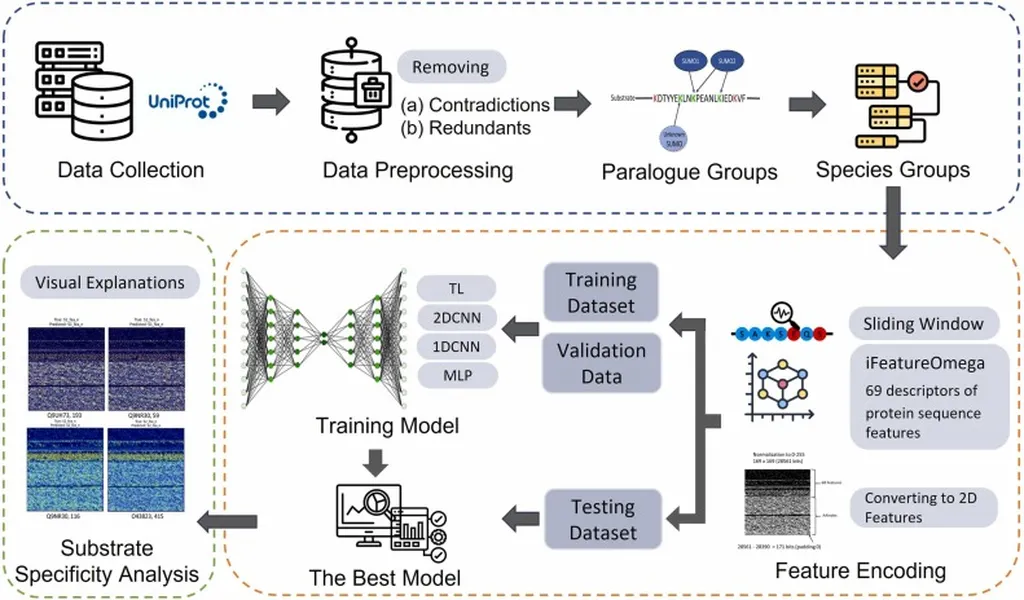
Enter SUMO-LMNet, a novel approach that integrates a lossless mapping strategy with deep learning architectures. “Our model extracts high-dimensional features from sequences and transforms them into two-dimensional feature maps, enabling convolutional neural networks (CNNs) to effectively capture both local and global dependencies within the data,” explains lead author Cheng-Hsun Ho from the Department of Medical Laboratory Science at I-Shou University in Kaohsiung City, Taiwan.
The significance of this research extends beyond the laboratory. In agriculture, understanding SUMOylation can lead to the development of crops with enhanced stress tolerance, improved yield, and better disease resistance. By accurately predicting SUMOylation sites, researchers can identify potential targets for genetic modification, paving the way for more resilient and productive agricultural systems.
SUMO-LMNet’s innovative approach includes a Lossless Mapping Network (LM-Net), which preserves the original feature space, ensuring that feature integrity is retained without loss of spatial information. This is a game-changer for the field, as it allows for more accurate and interpretable predictions.
The model’s effectiveness is underscored by its ability to achieve over 80% accuracy in distinguishing SUMO1 and SUMO2 modification sites. This high level of precision is crucial for prioritizing candidate sites for further study, accelerating the discovery of biologically relevant SUMOylation targets.
Moreover, the researchers introduced Combined Heatmap Feature Analysis (CHFA), which systematically aggregates feature importance across multiple samples, providing a more reliable and interpretable dataset-wide assessment. This method addresses the limitations of previous techniques like Grad-CAM, which lacked consistency across samples and did not offer a comprehensive evaluation of feature importance.
The implications of this research are vast. As we delve deeper into the complexities of gene regulation and protein function, tools like SUMO-LMNet will be instrumental in unraveling the intricacies of SUMOylation. This, in turn, can drive advancements in various fields, including agriculture, where the development of robust and high-yielding crops is a perpetual goal.
By making SUMO-LMNet publicly available, the researchers have opened the door for further exploration and collaboration. As Cheng-Hsun Ho notes, “Our model aids experimental design and accelerates the discovery of biologically relevant SUMOylation targets.” This open-access approach fosters a collaborative environment, where scientists can build upon existing knowledge and push the boundaries of what is possible.
In the realm of computational biology, SUMO-LMNet stands as a testament to the power of integrating deep learning with innovative strategies. As we continue to explore the vast potential of SUMOylation, tools like SUMO-LMNet will undoubtedly play a pivotal role in shaping the future of biological research and its applications in agriculture and beyond.