In the relentless battle against Verticillium wilt, a disease that threatens global cotton production, scientists have taken a significant step forward. A recent study published in *Industrial Crops and Products* has unveiled the intricate landscape of Nucleotide-binding domain and Leucine-rich Repeat (NLR) genes in cotton, offering new insights into how these genes respond to Verticillium dahliae infection. The research, led by Enhui Shen from the Institute of Digital Agriculture at the Zhejiang Academy of Agricultural Sciences and Zhejiang University, could pave the way for more resilient cotton varieties and bolster the agricultural sector against this devastating pathogen.
NLR proteins are crucial players in a plant’s immune system, acting as sentinels that recognize and respond to pathogen effectors. However, the comprehensive landscape of NLR responses to Vd-infection, particularly across different cotton cultivars, has remained largely unexplored—until now. Shen and his team identified 511 NLRs in upland cotton (Gossypium hirsutum L.) that were transcriptionally active in both resistant (MCU5) and susceptible (Siokra1–4) cultivars. Among these, 207 were supported by full-length transcripts obtained via third-generation long-read RNA-seq, revealing a complex tapestry of immune responses.
One of the most striking findings was the presence of cultivar-specific NLRs and alternative splicing events. “While many full-length NLRs were shared between MCU5 and Siokra1–4, a substantial number were cultivar-specific,” Shen explained. This suggests that different cotton varieties may employ unique transcriptional and post-transcriptional strategies to combat Vd-infection, a discovery that could have profound implications for breeding programs.
The study also delved into the expression patterns of homoeologous NLR pairs, revealing that At homoeologs were under relaxing purifying selection, while their corresponding Dt homoeologs were more conserved and showed higher expression. This balance between rapid immune response and evolutionary adaptability highlights the intricate mechanisms underlying cotton’s defense strategies.
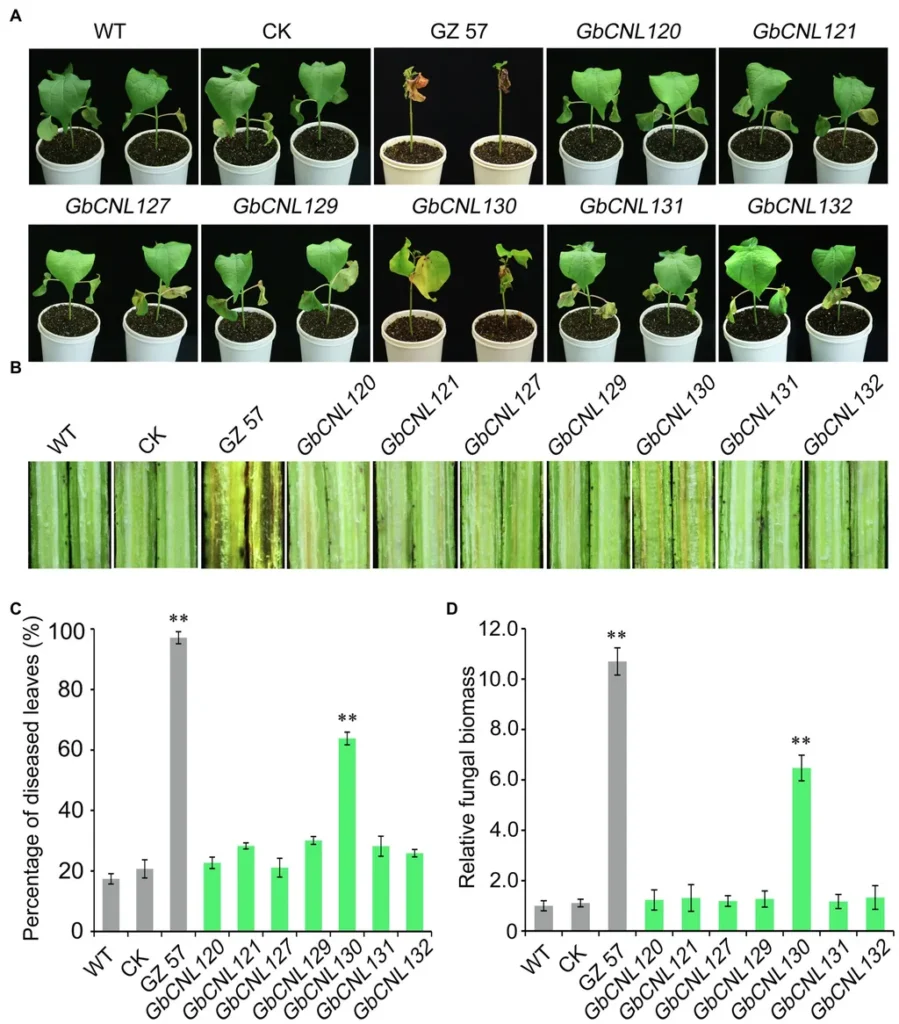
Functional validation further underscored the importance of NLRs in Vd-resistance. Silencing two specific NLRs significantly increased disease severity, demonstrating their positive role in basal resistance. “NLRs might contribute more to basal resistance in MCU5 than in Siokra1–4,” Shen noted, pointing to the potential for leveraging these genes to enhance disease resistance in susceptible varieties.
The commercial impacts of this research are substantial. By understanding the transcriptional landscape of NLRs, breeders can develop cotton varieties with improved resistance to Verticillium wilt, reducing crop losses and stabilizing yields. This is particularly crucial in regions where Vd-infection is rampant, threatening both small-scale farmers and large-scale agricultural operations.
Looking ahead, this work provides a valuable resource for further functional and breeding studies. As Enhui Shen and his team continue to unravel the complexities of cotton’s immune system, the agricultural sector stands to benefit from more resilient crops and innovative strategies to combat pathogens. The future of cotton farming is looking brighter, one NLR at a time.