In a significant stride towards revolutionizing agricultural diagnostics, researchers have developed a machine learning-driven system capable of rapidly identifying soilborne fungi, a critical factor in agricultural productivity and soil health. The study, published in *Applied Sciences*, introduces an automated imaging technique that promises to streamline the identification process, potentially saving farmers time and resources.
Soilborne fungi such as *Fusarium*, *Trichoderma*, *Verticillium*, and *Purpureocillium* play pivotal roles in disease dynamics and soil health. Traditional identification methods, however, are labor-intensive and time-consuming, often requiring up to 10 days for results. This delay can hinder timely interventions, impacting crop yields and overall farm management.
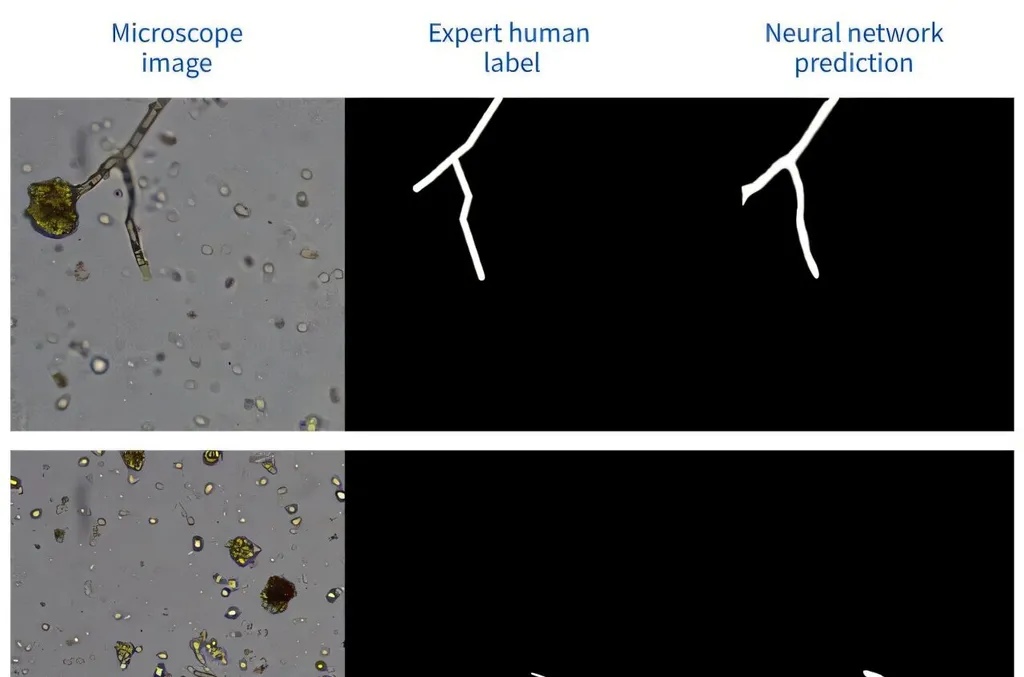
The research, led by Karol Struniawski from the Institute of Information Technology at Warsaw University of Life Sciences-SGGW, addresses these challenges by integrating robotic microscopy with deep learning. The team created the Soil Fungi Microscopic Images Dataset (SFMID), the largest publicly available dataset of its kind, comprising 20,151 images. This dataset includes both no-water and water-based images, providing a comprehensive resource for future research and development.
“We aimed to develop a system that could automate the identification process, making it faster and more accessible,” said Struniawski. “The integration of machine learning and automated microscopy allows for real-time inference, which is crucial for precision agriculture.”
The study evaluated four Convolutional Neural Network (CNN) architectures—InceptionResNetV2, ResNet152V2, DenseNet121, and DenseNet201—using transfer learning and three-shot majority voting. The ResNet152V2 conv2 architecture achieved optimal performance with a precision of 0.6711 and an AUC of 0.8031, demonstrating the system’s efficacy in identifying soil fungi.
Grad-CAM analysis was employed to validate the biological relevance of the model’s predictions, ensuring that the machine learning algorithms were focusing on the correct features. Statistical validation through McNemar’s test confirmed that three-shot classification significantly outperforms single-image prediction, highlighting the importance of multiple images in accurate identification.
The research also identified challenges in distinguishing between certain fungi, such as *Fusarium* and *Trichoderma* in no-water conditions, and *Fusarium* and *Verticillium* in water-based conditions. These findings point to morphological ambiguities that could be addressed in future research.
The commercial implications of this research are substantial. By providing a scalable and publicly available dataset, the study lays the groundwork for AI-enhanced agricultural diagnostics. Farmers and agronomists can benefit from faster, more accurate identification of soilborne fungi, enabling timely interventions and improved crop management.
“This research not only advances the field of agricultural diagnostics but also sets a new standard for the integration of machine learning and automated microscopy,” said Struniawski. “The potential applications are vast, and we are excited to see how this technology will be adopted and further developed in the future.”
As the agricultural sector continues to embrace precision farming techniques, the development of automated, AI-driven diagnostic tools will be crucial. This research, published in *Applied Sciences* and led by Karol Struniawski from the Institute of Information Technology at Warsaw University of Life Sciences-SGGW, represents a significant step forward in this direction, offering a glimpse into the future of agricultural technology.