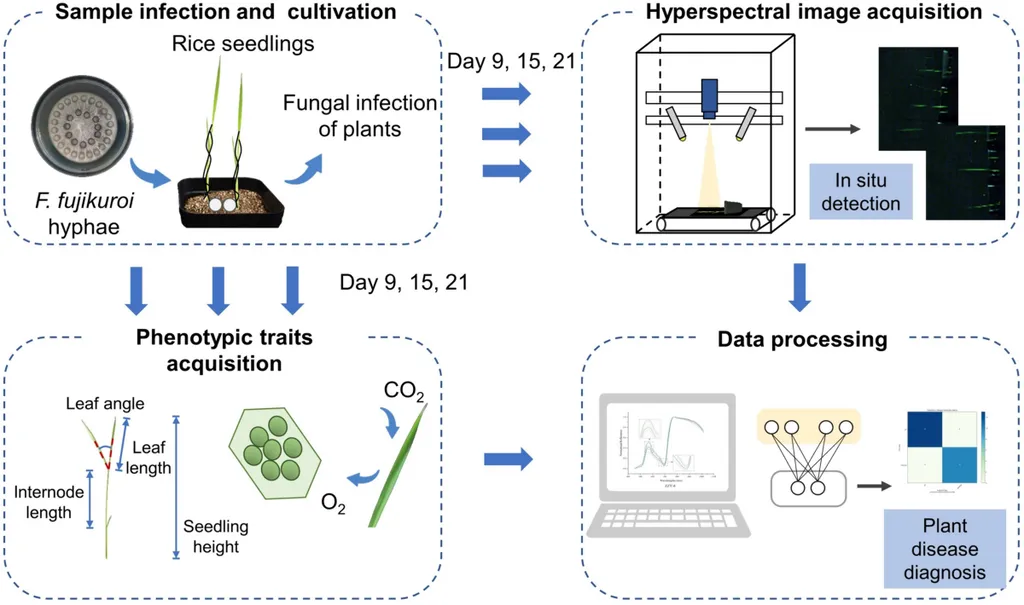
In the heart of China’s Zhejiang province, a groundbreaking study is set to revolutionize the way we diagnose and combat one of rice’s most formidable foes: bakanae disease. This fungal menace, caused by Fusarium fujikuroi, has long threatened global rice production, causing severe yield losses and sending shockwaves through the agriculture sector. But now, a team of researchers led by Sishi Chen from the College of Biosystems Engineering and Food Science at Zhejiang University has integrated the power of the plant microbiome with machine learning (ML) to develop a precision diagnosis tool that could change the game.
The study, published in *Plant Methods*, delves into the intricate world of the plant microbiome, a complex ecosystem that plays a pivotal role in plant stress resistance. “The microbiome is like an invisible shield, protecting plants from diseases,” explains Chen. “But its high-dimensional characteristics have not been fully exploited—until now.”
The researchers found significant correlations between certain bacterial groups, specifically Gammaproteobacteria and Bacteroidia, and the severity of bakanae disease. This discovery opened the door to developing advanced diagnostic models. The team constructed different models based on random forests (RF), support vector machines (SVM), and convolutional neural networks (CNN) using data from 88 biological replicates.
The results were impressive. The RF model demonstrated strong performance across four taxonomic levels, with an accuracy of 88.9% and an F1 score of 94.1%. “This is a significant leap forward,” says Chen. “The RF model’s ability to handle high-dimensional data makes it particularly suitable for this kind of analysis.”
But the innovation doesn’t stop there. The researchers proposed a Bray-Curtis dissimilarity-based extraction method to rapidly screen practical information from the original microbial community. This method enhances model performance, making the diagnostic process more efficient and accurate.
The study also classified the disease severity of infected samples into two levels (high and low infected levels) using the K-means clustering method. In diagnosing infection severity based on the family level, the model’s prediction accuracy reached 77.8%. “This level of precision is crucial for farmers,” Chen notes. “It allows for targeted interventions, reducing the need for broad-spectrum fungicides and minimizing environmental impact.”
The commercial implications of this research are vast. Precision diagnosis tools can help farmers identify and treat diseased plants more effectively, reducing yield losses and increasing profitability. “This technology has the potential to transform the agriculture sector,” says Chen. “By integrating microbiome data with machine learning, we can develop more precise, efficient, and sustainable agricultural practices.”
The study’s findings highlight the potential of combining microbiome research with advanced technologies like machine learning. This integration could pave the way for new diagnostic strategies and precision agriculture tools, ultimately benefiting farmers and the global food supply.
As we look to the future, the fusion of microbiome research and machine learning holds immense promise. It’s a testament to the power of interdisciplinary collaboration and the potential of technology to address some of the most pressing challenges in agriculture. With further research and development, this approach could be applied to other crops and diseases, opening up new avenues for innovation and growth in the agriculture sector.