In the heart of Pakistan, researchers have uncovered a genetic treasure trove that could revolutionize cotton farming, particularly in the face of climate change. Muhammad Tayyab, a scientist at the Centre of Agricultural Biochemistry and Biotechnology at the University of Agriculture Faisalabad, has led a groundbreaking study that could pave the way for more resilient cotton crops.
The study, published in the Journal of Cotton Research (which translates to “Journal of Cotton Research” in English), delves into the world of bromodomain (BRD) proteins, which play a crucial role in regulating gene expression. These proteins, known for recognizing acetylated lysine residues, act as chromatin-associated post-translational modification-inducing proteins. While their roles in mammals and model plants like Arabidopsis and rice have been extensively studied, their functions in cotton species have remained a mystery—until now.
Tayyab and his team identified 145 BRD genes in tetraploid cotton species (Gossypium hirsutum and G. barbadense), compared to 82 in their diploid progenitors (G. arboreum and G. raimondii). This significant difference highlights the impact of polyploidization on BRD gene evolution. “The expansion of BRD genes through segmental and tandem duplications suggests a complex evolutionary history,” Tayyab explains. “This expansion could be a key factor in the cotton plant’s ability to adapt to various environmental stresses.”
The researchers found that most BRD genes were intron-less and conserved throughout evolution, indicating their fundamental role in cotton’s genetic makeup. Syntenic analysis revealed a greater number of orthologous gene pairs in the Dt sub-genome than in the At sub-genome, suggesting a potential functional divergence between the two sub-genomes.
One of the most compelling findings was the abundance of regulatory, hormonal, and defense-related cis-regulatory elements in the promoter region of BRD genes. This suggests that BRD genes play a significant role in both biotic and abiotic stress responses. Protein-protein interaction analysis indicated that global transcription factor group E (GTE) transcription factors regulate BRD genes, further underscoring their importance in stress tolerance.
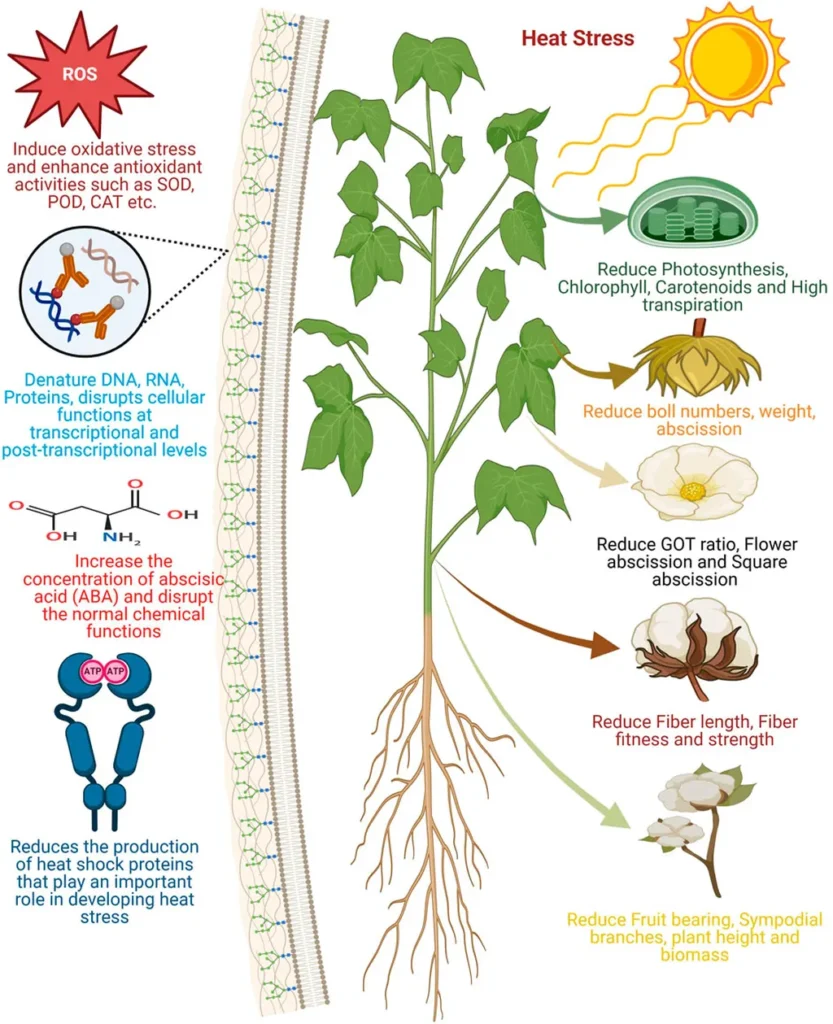
Expression analysis revealed that BRD genes are predominantly involved in ovule development, with some genes displaying specific expression patterns under heat, cold, and salt stress. “The differential expression of BRD genes between tolerant and sensitive genotypes underscores their potential role in mediating drought and salinity stress responses,” Tayyab notes. This finding could be a game-changer for cotton farmers, particularly those in regions prone to drought and salinity.
The implications of this research extend beyond the agricultural sector. As climate change continues to pose challenges to global food security, the development of stress-tolerant crops becomes increasingly important. The insights gained from this study could shape future breeding programs, leading to the development of cotton varieties that are more resilient to environmental stresses.
Moreover, the understanding of BRD genes’ roles in stress tolerance could have broader applications in the energy sector. Cotton is not only a vital crop for textiles but also a source of biofuel. Developing stress-tolerant cotton varieties could enhance biofuel production, contributing to a more sustainable energy future.
In conclusion, Tayyab’s research provides valuable insights into the evolution of BRD genes across species and their roles in abiotic stress tolerance. As we face the challenges of a changing climate, this study offers a beacon of hope for more resilient crops and a more sustainable future. The findings, published in the Journal of Cotton Research, mark a significant step forward in our understanding of cotton genetics and its potential applications in agriculture and beyond.