In the relentless battle against antimicrobial resistance, scientists are uncovering the intricate dance between resistance mechanisms, genetic variations, and virulence factors in some of the most troublesome pathogens. A recent study published in *The Microbe* (translated to English as *The Microbe*) sheds light on the complex interplay of polymyxin resistance in Escherichia coli and Klebsiella pneumoniae, offering critical insights that could reshape our approach to combating these superbugs.
Led by Anelise Stella Ballaben from the Department of Agricultural and Environmental Biotechnology at São Paulo State University (UNESP) and the School of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo, the research delves into the phenotypic and genotypic features of polymyxin-resistant clinical isolates from Brazilian hospitals. The study analyzed 17 Gram-negative bacilli, including 14 E. coli and 3 K. pneumoniae strains, using a combination of antimicrobial susceptibility testing, lipid A profiling via mass spectrometry, whole-genome sequencing (WGS), and virulence assessment through the Galleria mellonella infection model.
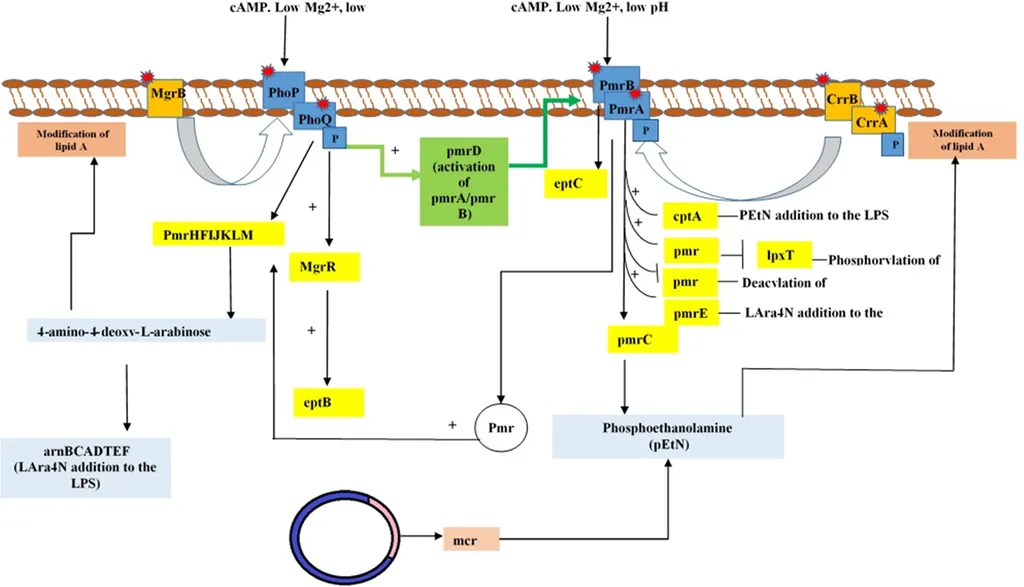
The findings reveal a sophisticated web of resistance mechanisms. “We observed structural modifications of lipid A, such as the addition of phosphoethanolamine (PEtN), L-Ara4N, palmitate, and LpxO-mediated hydroxylation, which significantly alter the bacterial cell surface and reduce polymyxin binding,” explains Ballaben. These modifications, coupled with the presence of major resistance genes like mcr-1.1, blaNDM-1, and blaCTX-M-15, highlight the adaptability of these pathogens.
The genetic context analysis further uncovered associations with mobile elements like ISEcp1 and ISCR1, as well as integron-associated gene cassettes (e.g., aadA2, dfrA12). “The mobility of these genetic elements facilitates the rapid dissemination of resistance genes, making it a formidable challenge for clinicians and public health officials,” notes Ballaben.
The study also revealed high clonal diversity among the E. coli isolates, including sequence types (STs) like ST10, ST131, ST354, and ST410, and the detection of K. pneumoniae ST11 and ST4477. Notably, ST11 is a dominant high-risk clone both in Brazil and globally, underscoring the widespread nature of these resistant strains.
Virulence profiling added another layer of complexity, with some strains exhibiting hypervirulent phenotypes. “The heterogeneous nature of these virulence profiles suggests that resistance and virulence are not mutually exclusive but can coexist, posing a dual threat to patient outcomes,” Ballaben adds.
The implications of this research extend beyond clinical settings. Understanding the genetic and phenotypic landscape of these pathogens is crucial for developing targeted therapies and surveillance strategies. For the energy sector, where microbial contamination can impact operations, particularly in water treatment and biofuel production, these findings could inform better biosecurity measures and antimicrobial stewardship programs.
As Ballaben concludes, “Our study underscores the importance of genomic surveillance to monitor the dissemination of multidrug-resistant (MDR) and extensively drug-resistant (XDR) pathogens. This knowledge is vital for developing effective interventions and mitigating the public health impact of these superbugs.”
With the insights gleaned from this research, the scientific community is better equipped to tackle the evolving challenge of antimicrobial resistance, ensuring that our last-resort antibiotics remain effective for as long as possible.