In a groundbreaking development that could revolutionize the field of antimicrobial research, scientists have harnessed the power of deep learning to design highly effective antimicrobial peptides (AMPs). This innovative approach, detailed in a study published in *Nature Communications* (translated as “Nature Communications”), promises to accelerate the discovery of new bioactive peptides, addressing critical challenges in healthcare and agriculture.
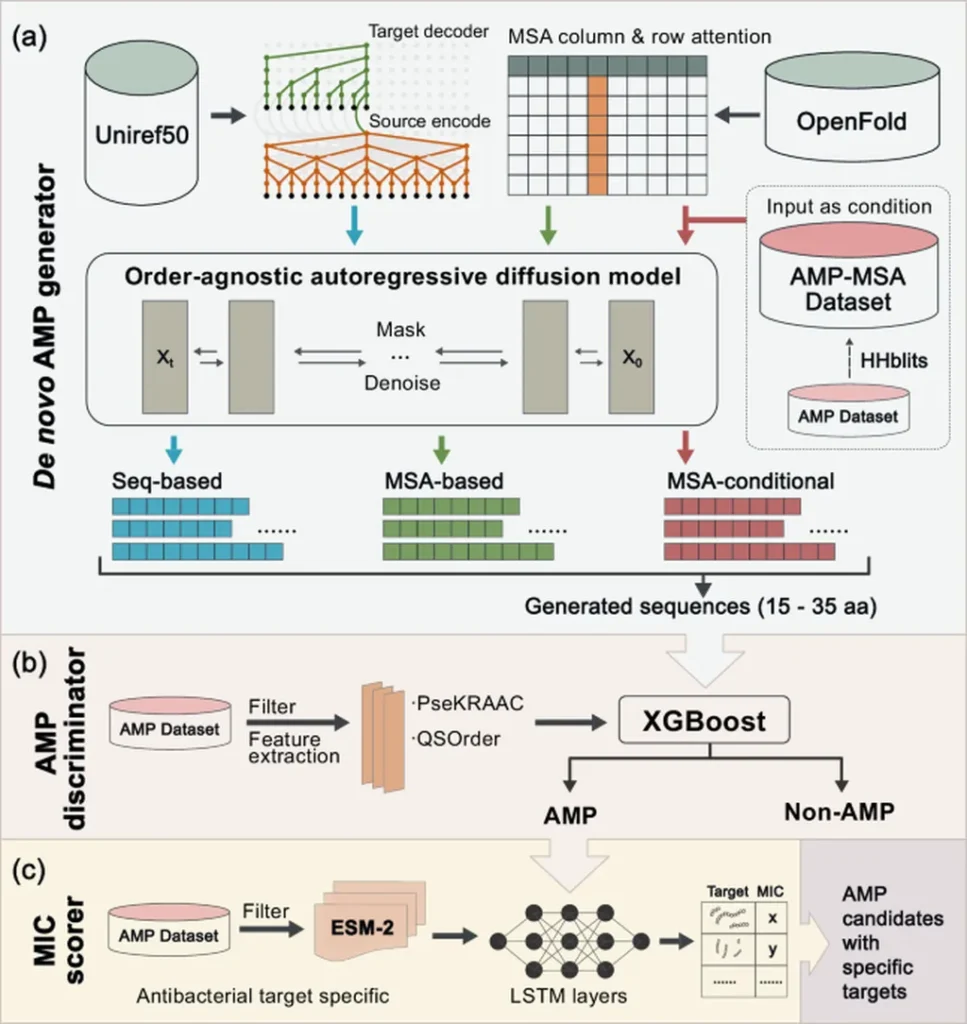
The research, led by Han Gao from the State Key Laboratory of Animal Nutrition and Feeding at the Institute of Animal Science, Chinese Academy of Agricultural Sciences, introduces a novel strategy called DLFea4AMPGen. This method leverages deep learning models to identify and extract key features associated with antimicrobial activity, enabling the generation of peptide sequences with potential bioactivities.
“Current methods for designing antimicrobial peptides often face challenges such as low success rates and the need to screen through large virtual libraries,” explains Han Gao. “Our approach streamlines this process by focusing on the most significant features, making it more efficient and effective.”
The study employed the SHapley Additive exPlanations (SHAP) method to quantify the contribution of each amino acid in multifunctional peptides, which exhibit antibacterial, antifungal, and antioxidant activities. By extracting key feature fragments (KFFs) with the highest average contributions, the researchers classified these fragments into four subfamilies based on amino acid frequency. This systematic arrangement generated a plausible sequence subspace for candidate peptides.
From this subspace, 16 representative sequences were selected for experimental validation. Impressively, 75% (12/16) of these sequences demonstrated at least two types of activity. Notably, one of the peptides, designated D1, exhibited broad-spectrum antimicrobial activity, including efficacy against multidrug-resistant clinical pathogenic isolates both in vitro and in vivo.
“This proof-of-concept study underscores the potential of the DLFea4AMPGen platform for efficient design and screening of bioactive peptides,” says Han Gao. “It showcases the value of integrating deep learning models in AMP research, offering a powerful tool for future developments in the field.”
The implications of this research are far-reaching. By accelerating the discovery and optimization of antimicrobial peptides, DLFea4AMPGen could significantly impact the healthcare sector, particularly in the fight against antibiotic-resistant infections. Additionally, the agricultural industry stands to benefit from the development of new antimicrobial agents to combat plant pathogens and improve crop yields.
As the world grapples with the growing threat of antimicrobial resistance, innovative solutions like DLFea4AMPGen offer a beacon of hope. By leveraging the power of artificial intelligence and deep learning, researchers are paving the way for a new era of antimicrobial discovery, one that promises to enhance our ability to combat infectious diseases and safeguard global health.
In the words of Han Gao, “This research represents a significant step forward in our quest to develop more effective and efficient antimicrobial agents. The potential applications are vast, and we are excited to explore the many possibilities that this technology offers.”