In the world of crop science, understanding the genetic diversity of our food plants is crucial for improving yields, resilience, and quality. A recent study published in *Frontiers in Plant Science* (translated to *Frontiers in Plant Science*) has shed new light on the genetic divergence between Eastern and Western gene pools of carrot (Daucus carota L.), offering insights that could revolutionize carrot breeding programs and potentially benefit the agricultural sector.
The study, led by Trupthi Mudihal from the Plant Molecular Biology Lab at the University of Horticultural Sciences in Bagalkot, Karnataka, India, is the first to integrate agro-morphometric traits with functional InDel (Insertion-Deletion) markers to unravel the genetic differences between these two gene pools. “Our research aimed to bridge the gap between phenotypic diversity and genetic variation in carrots,” Mudihal explained. “By identifying functional InDels, we hoped to provide a tool for breeders to develop improved carrot cultivars adapted to diverse climates.”
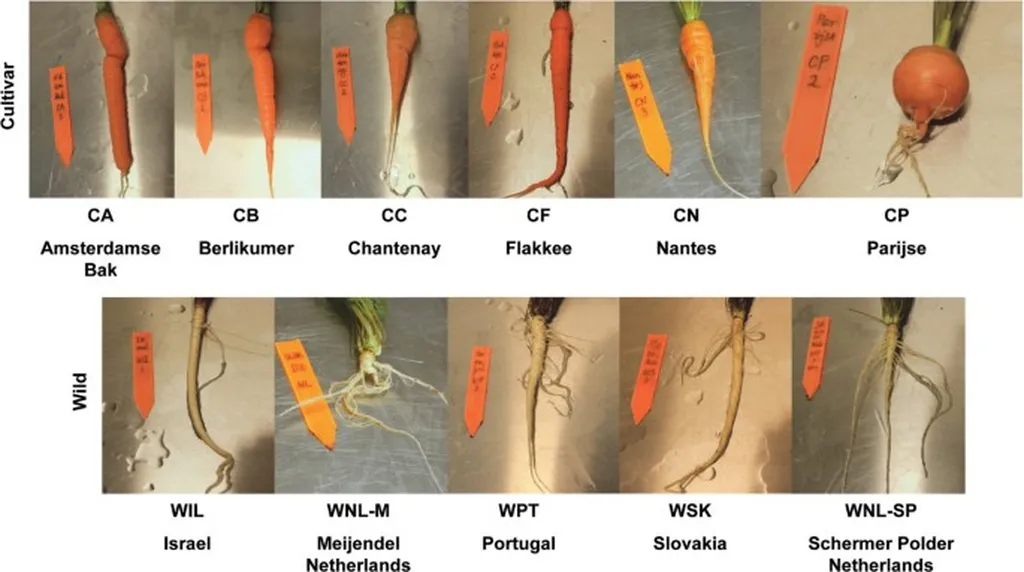
Carrots are a globally cultivated root vegetable, and their genetic diversity is significant. The study found that Eastern carrot accessions exhibited larger plants with bigger, more colorful roots, while Western accessions were more uniform in color and compact in architecture. This divergence is not just skin deep; it reflects underlying genetic differences that could be harnessed for breeding programs.
From RNA-seq data, the researchers identified 271 agarose-resolvable functional InDels, with a length difference of more than 15 base pairs. Of these, 48 validated markers showed high polymorphism (84.21%) across the two gene pools, supporting secondary domestication changes. These InDels, located in coding and untranslated regions (UTRs), likely regulate gene expression and may have contributed to significant genetic modifications among carrot gene pools.
The study also revealed that the Western gene pool exhibited more intense selection and domestication, suggesting a history of targeted breeding for specific traits. Population structure and phylogenetic analysis revealed clear gene pool differentiation (Fst = 0.181) with potential gene flow (Nm = 1.716). Functional annotation of linked InDels to key biological processes highlighted their role in domestication.
Key InDels such as DcFInDel32, DcFInDel28, and DcFInDel55 were associated with multiple traits, underscoring their utility in marker-assisted selection (MAS). “These findings provide a roadmap for developing improved carrot cultivars with high yield and quality,” Mudihal noted. “They also offer a deeper understanding of the genetic basis of important traits, which can be leveraged for future breeding programs.”
The implications of this research extend beyond the carrot patch. By understanding the genetic diversity and domestication history of carrots, breeders can develop cultivars that are more resilient to climate change, have higher yields, and are better adapted to local conditions. This could lead to more sustainable and productive agricultural practices, benefiting both farmers and consumers.
Moreover, the study’s findings could have broader applications in the agricultural sector, particularly in the development of crops that are more resistant to pests, diseases, and environmental stresses. As the global population continues to grow, the demand for food will only increase, making it more important than ever to develop crops that can meet this demand sustainably.
In conclusion, this research represents a significant step forward in our understanding of carrot genetics and has the potential to shape the future of carrot breeding programs. As Mudihal and her team continue to explore the genetic diversity of carrots, they are paving the way for a more sustainable and productive future for this important crop. The study was published in *Frontiers in Plant Science*, a leading journal in the field of plant science, and is available for further reading.