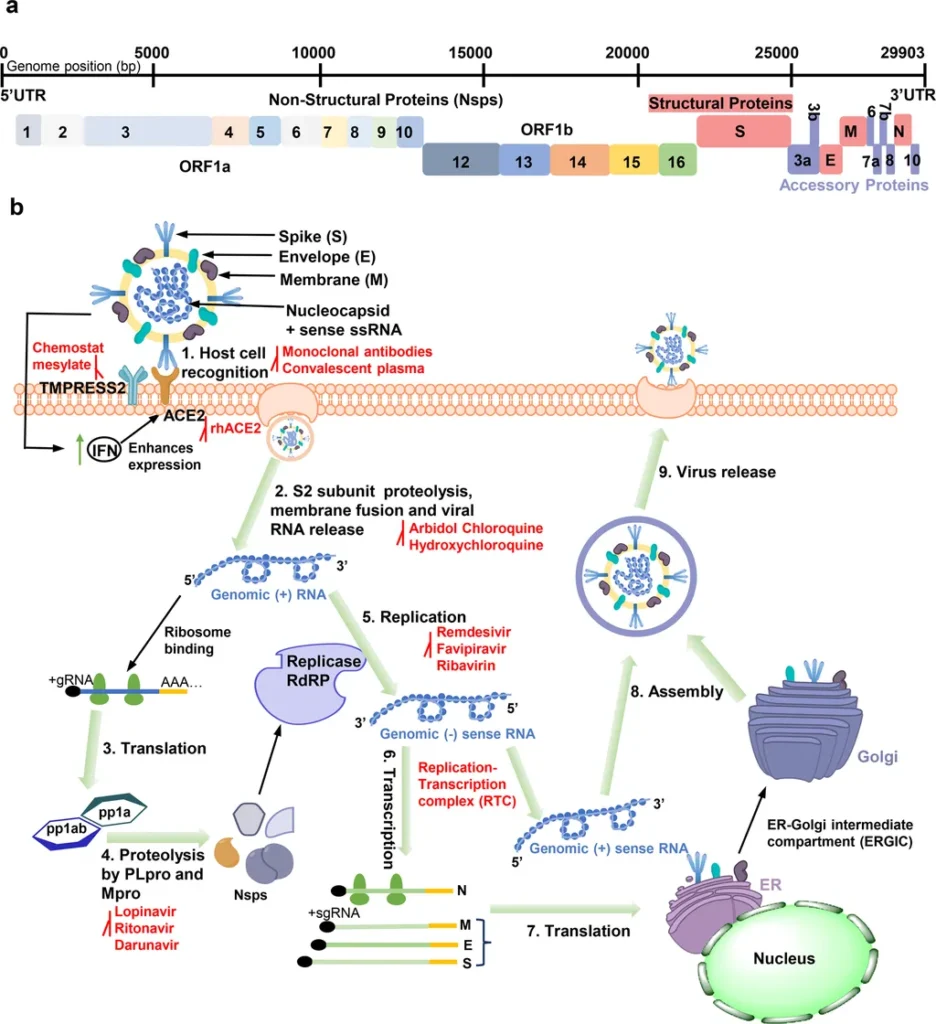
In the relentless pursuit to understand and combat COVID-19, researchers have turned to the intricate world of protein structures, seeking insights that could unlock new treatments and strategies. A recent study published in *Discover Applied Sciences* has taken a significant step in this direction by conducting a comparative structural analysis of key host proteins involved in SARS-CoV-2 infection. The research, led by Ahmed S. Fadel from the Department of Computer Science at Ahram Canadian University, sheds light on the structural similarities between these proteins and others, potentially paving the way for drug repurposing and the design of targeted inhibitors.
The study focuses on three critical human proteins: Angiotensin-converting enzyme 2 (ACE2), Transmembrane protease serine 2 (TMPRSS2), and Furin. These proteins play pivotal roles in the pathogenesis of SARS-CoV-2 and the viral entry into host cells. By leveraging 3D structure data from the SCOP and PDB databases, the research team performed structural alignments of these proteins using various algorithms. The goal was to identify conserved binding pockets among structurally similar proteins, which could serve as targets for therapeutic intervention.
The findings revealed notable similarities. For instance, the ACE2 protein showed the highest similarity to Thimet oligopeptidase, a proteolysis protein. TMPRSS2, on the other hand, exhibited a striking similarity to Enteropeptidase-1, with a similarity score of 0.94. Furin, another crucial protein in viral entry, was found to be highly similar to Kexin, a hydrolase/hydrolase inhibitor protein.
These structural insights are not just academic exercises; they hold significant promise for practical applications. “The structural similarities we identified offer critical pathways for repurposing existing drugs and designing new inhibitors targeting ACE2, TMPRSS2, and Furin,” explained Fadel. “This could aid not only in the fight against COVID-19 but also in preparing for future viral infections.”
The implications for the agriculture sector are particularly noteworthy. Understanding the structural similarities between viral and host proteins can lead to the development of more effective and targeted treatments for livestock and crops affected by similar viral pathogens. This could result in reduced losses and increased productivity, benefiting farmers and the agricultural industry as a whole.
Moreover, the study highlights the importance of bioinformatics and structural biology in drug discovery and repurposing. By identifying conserved binding pockets, researchers can accelerate the development of new therapies, potentially reducing the time and cost associated with traditional drug discovery processes.
As the world continues to grapple with the COVID-19 pandemic, research like this offers a glimmer of hope. It underscores the power of interdisciplinary approaches in tackling complex health challenges and opens new avenues for innovation in both the medical and agricultural sectors. The study, published in *Discover Applied Sciences* and led by Ahmed S. Fadel from the Department of Computer Science at Ahram Canadian University, stands as a testament to the potential of structural biology in shaping the future of healthcare and agriculture.