In the heart of agricultural innovation, a groundbreaking study has unveiled the intricate workings of the Cytochrome P450 supergene family in citrus species, offering promising insights for the future of citrus cultivation, particularly under stress conditions. Published in *BMC Plant Biology*, this research, led by Arfa Tanveer from the Center of Agricultural Biotechnology and Biochemistry at the University of Agriculture Faisalabad, delves into the genetic mechanisms that could revolutionize how we approach citrus farming.
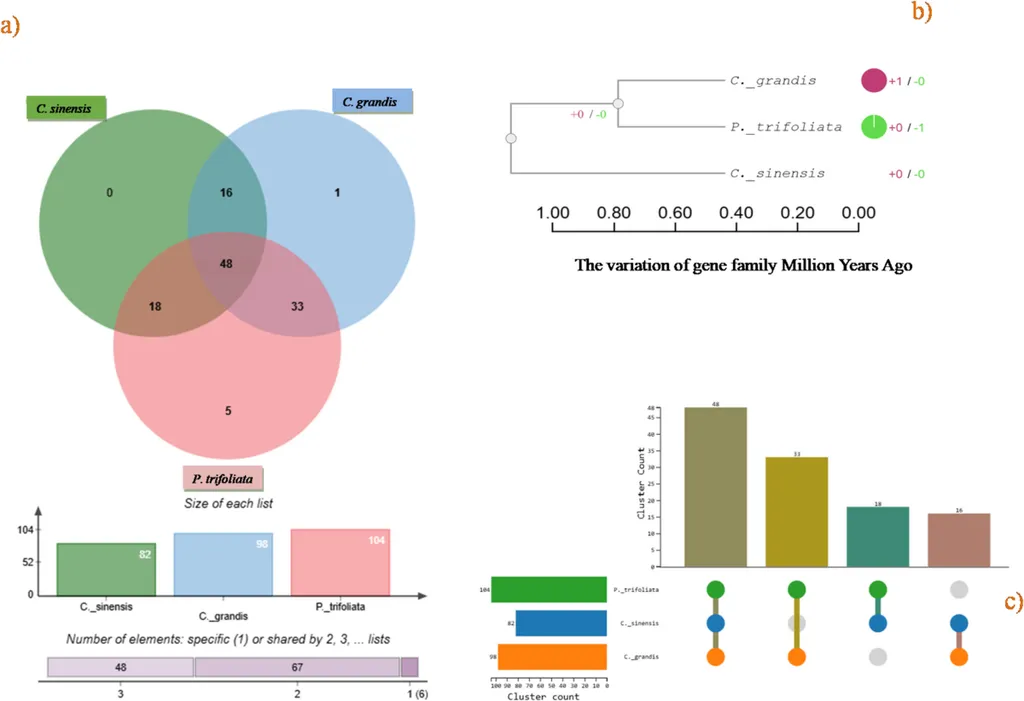
The study focuses on three citrus species: *C. sinensis* (sweet orange), *C. grandis* (pomelo), and *P. trifoliata* (trifoliate orange). By conducting a genome-wide analysis, the researchers identified 510 members of the P450 gene family, which play a crucial role in plant growth, development, and biosynthesis. These genes vary significantly in size, molecular weight, and isoelectric points, highlighting the diversity within this supergene family.
One of the most compelling findings is the phylogenetic classification of these genes into 43 subfamilies, further divided into clans. Notably, *C. sinensis* and *C. grandis* each have 8 clans, while *P. trifoliata* has 6. This classification provides a deeper understanding of the evolutionary relationships and functional diversity within the P450 gene family.
The study also revealed that these genes are distributed across various organelles, indicating their multifunctional roles within the plant. The presence of conserved motifs and varying numbers of introns and exons among species suggests a complex regulatory mechanism that could be harnessed for agricultural benefits.
Arfa Tanveer emphasized the practical implications of these findings: “Understanding the regulatory elements and expression patterns of these genes under different stress conditions can help us develop more resilient citrus varieties. This is particularly important for regions facing increasing abiotic stresses like heat and drought.”
The synteny analysis revealed divergence and col-linearity relationships among the genes of the three species, providing insights into their evolutionary history and potential functional similarities. The protein-protein interaction (PPI) network analysis further showed strong interactions among all CYPs, suggesting a highly interconnected regulatory network.
Expression analysis under biotic and abiotic stress conditions revealed significant variations in gene expression, with more pronounced changes observed under abiotic stresses. This finding is crucial for developing strategies to mitigate the impact of environmental stresses on citrus crops.
The commercial implications of this research are vast. By identifying key genes involved in stress responses, breeders can develop citrus varieties that are more resilient to environmental challenges. This could lead to increased yields, improved quality, and greater economic returns for farmers.
As the agricultural sector faces increasing pressures from climate change and environmental stresses, this research offers a beacon of hope. The insights gained from this study could pave the way for innovative breeding programs and biotechnological interventions that enhance the resilience and productivity of citrus crops.
In the words of Arfa Tanveer, “This research not only advances our understanding of the P450 gene family but also opens new avenues for improving citrus cultivation. The potential to develop stress-resistant varieties is a game-changer for the agricultural sector.”
As we look to the future, the findings from this study could shape the next generation of citrus farming, ensuring that this vital crop continues to thrive in the face of evolving environmental challenges.