In the ever-evolving landscape of genomic research, a groundbreaking study has emerged, promising to reshape our understanding of genetic linkages across species. Published in the prestigious journal *Genome Biology*, the research introduces X-LDR, a novel stochastic algorithm designed to map linkage disequilibrium (LD) on an unprecedented scale. This innovation could have profound implications for the agriculture sector, offering new tools to enhance crop breeding and livestock management.
Linkage disequilibrium refers to the non-random association of alleles at different loci in a given population. Understanding LD patterns is crucial for identifying genetic variations that contribute to traits of interest, such as disease resistance, yield, and adaptability in plants and animals. However, the sheer volume of genomic data has posed significant challenges in scaling LD analysis to the entire genome.
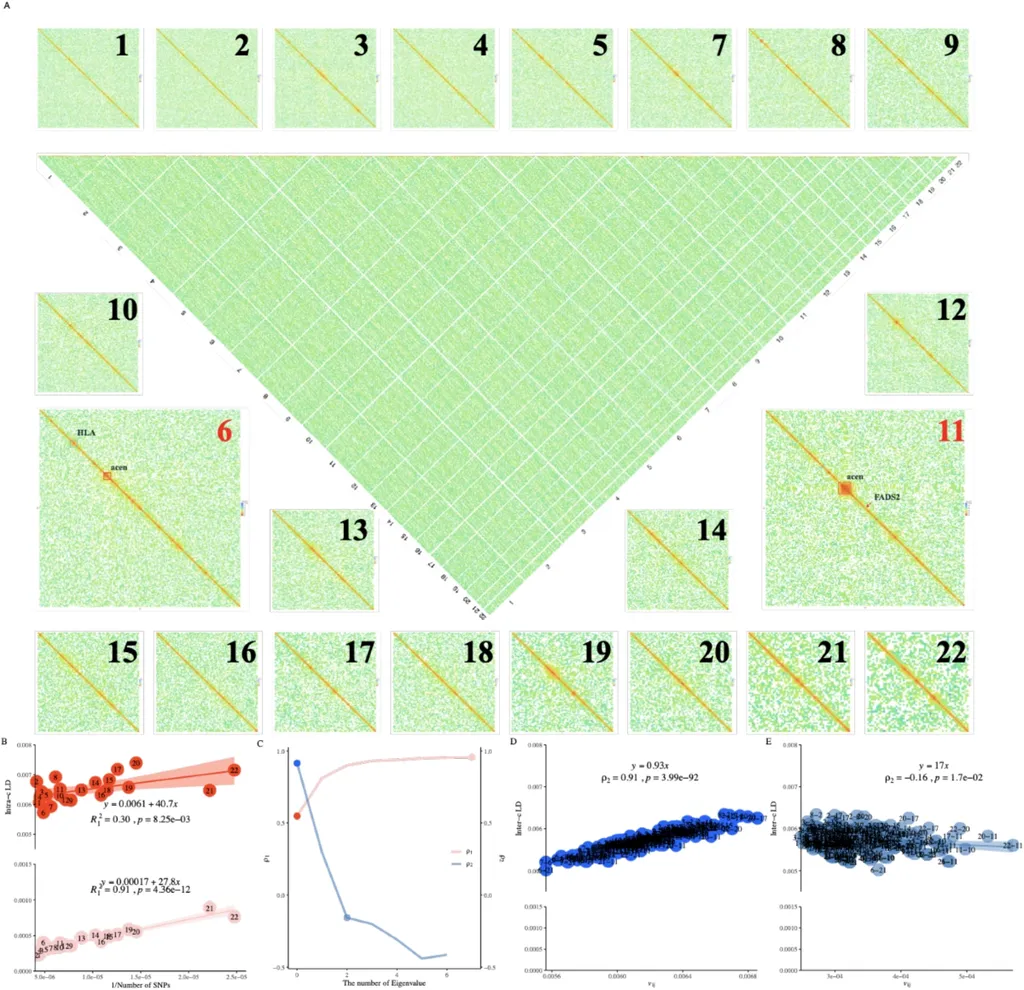
Enter X-LDR, a computational algorithm developed by lead author Tian-Neng Zhu and colleagues at the Department of Genetic and Genomic Medicine, Center for Laboratory Medicine, Zhejiang Provincial People’s Hospital. The algorithm is designed to handle biobank-scale data efficiently, scaling the entire genome to high-resolution LD grids. For instance, X-LDR can process nearly 9 million LD grids for the UK Biobank, which includes data from approximately 300,000 samples and 4.2 million single nucleotide polymorphisms (SNPs).
“The X-LDR algorithm represents a significant leap forward in our ability to analyze genetic linkages,” said Zhu. “Its efficiency and scalability allow us to uncover intricate patterns of LD that were previously inaccessible, providing a more comprehensive understanding of genetic diversity.”
The study not only scales LD across the human genome but also presents an unprecedented LD atlas for 25 reference populations, contouring the diversity of interspecies LD. This cross-species analysis could be a game-changer for agriculture, where understanding genetic linkages can lead to more effective breeding programs and improved crop and livestock varieties.
“In agriculture, the ability to map LD across different species can help identify genetic markers associated with desirable traits,” explained Zhu. “This can accelerate the development of new crop varieties that are more resilient to environmental stresses and diseases, ultimately enhancing food security.”
The commercial impacts of this research are substantial. Farmers and agricultural companies can leverage the insights gained from X-LDR to optimize breeding strategies, reduce costs, and increase yields. The algorithm’s efficiency also makes it a valuable tool for researchers and breeders working with large genomic datasets, enabling faster and more accurate genetic analyses.
Looking ahead, the X-LDR algorithm could pave the way for further advancements in genomic research. Its ability to handle vast amounts of data efficiently opens up new possibilities for studying genetic linkages in diverse species, from crops to livestock. This could lead to breakthroughs in understanding the genetic basis of complex traits and developing targeted breeding programs.
As the field of genomics continues to evolve, tools like X-LDR will be instrumental in unlocking the full potential of genetic data. The research published in *Genome Biology* by lead author Tian-Neng Zhu and his team at the Department of Genetic and Genomic Medicine, Center for Laboratory Medicine, Zhejiang Provincial People’s Hospital, marks a significant milestone in this journey, offering a glimpse into the future of genetic research and its transformative impact on agriculture.