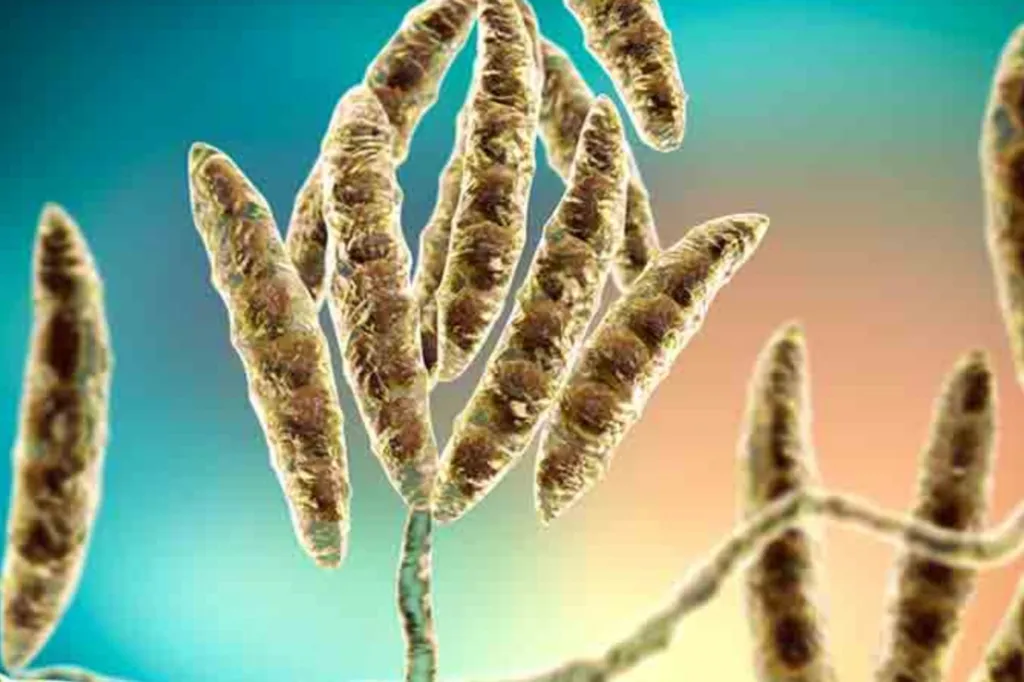
In the ever-evolving landscape of agricultural science, a recent study has shed new light on the Fusarium fujikuroi species complex (FFSC), a group of fungi with significant implications for global agriculture. The research, published in *The Plant Pathology Journal*, revisits the taxonomic identities of 81 Korean isolates within the FFSC, revealing critical insights that could reshape how we understand and manage these phytopathogenic and mycotoxigenic species.
The study, led by Le Dinh Thao of the Korean Agricultural Culture Collection, employed multi-locus sequence analyses to re-identify the isolates using partial gene fragments of five key genes: translation elongation factor 1-alpha (tef1), beta-tubulin (tub2), calmodulin (CaM), RNA polymerase II largest subunit (rpb1), and RNA polymerase II second largest subunit (rpb2). This meticulous approach clarified the taxonomic identities of these isolates, uncovering several surprises.
“Many strains previously reported as F. proliferatum, F. subglutinans, and F. circinatum were re-identified as F. annulatum, F. dendrobii, and a novel species, F. ipomoeicola sp. nov., respectively,” Thao explained. This revision not only adds a new species to the FFSC but also revises the taxonomic status of four recently described species, indicating that they are synonyms of previously known species.
The implications for the agriculture sector are substantial. Accurate identification of these fungal species is crucial for developing effective management strategies and preventing crop losses. The study confirmed the presence of eight species within the FFSC in Korea, including seven known species and one novel species. Notably, F. annulatum, F. dendrobii, F. elaeagni, and F. planum are reported for the first time in Korea, along with 22 previously undocumented fungus-host associations.
“This research highlights the importance of precise taxonomic identification in understanding the distribution and impact of these fungi,” said Thao. “It also underscores the need for continuous monitoring and revision of fungal species to keep pace with their evolutionary dynamics.”
The study’s findings could influence future developments in agricultural biotechnology and plant pathology. By providing a clearer picture of the FFSC’s taxonomic landscape, the research paves the way for more targeted and effective control measures. This could include the development of resistant crop varieties, improved diagnostic tools, and more effective fungicides.
Moreover, the identification of novel fungus-host associations opens new avenues for research into the ecological roles and economic impacts of these fungi. Understanding these interactions could lead to better integrated pest management strategies, ultimately benefiting farmers and the broader agricultural industry.
As the study concludes, the pathogenicity of these fungal species on their respective hosts was not confirmed, leaving room for further investigation. However, the groundwork laid by this research is invaluable, setting the stage for future studies that could unlock even greater insights into the complex world of plant pathogens.
In the dynamic field of agritech, this study serves as a reminder of the critical role that taxonomic revision plays in advancing our understanding of agricultural challenges. With the agricultural sector facing increasing pressures from climate change and global trade, accurate and up-to-date knowledge of plant pathogens is more important than ever.
As the research was published in *The Plant Pathology Journal*, led by Le Dinh Thao of the Korean Agricultural Culture Collection, Agricultural Microbiology Division, National Institute of Agricultural Sciences, Rural Development Administration, it underscores the global relevance and collaborative nature of agricultural research. The findings not only benefit Korea but also have the potential to inform and improve agricultural practices worldwide.