In the intricate world of plant microbiomes, a recent study has shed light on the niche-specific microbial diversity and interactions within spinach, offering valuable insights that could revolutionize agricultural practices. The research, published in *Current Research in Microbial Sciences*, delves into the complex relationships between spinach (Spinacia oleracea L.) and its associated microbial communities, highlighting the potential for targeted strategies to enhance crop health and productivity.
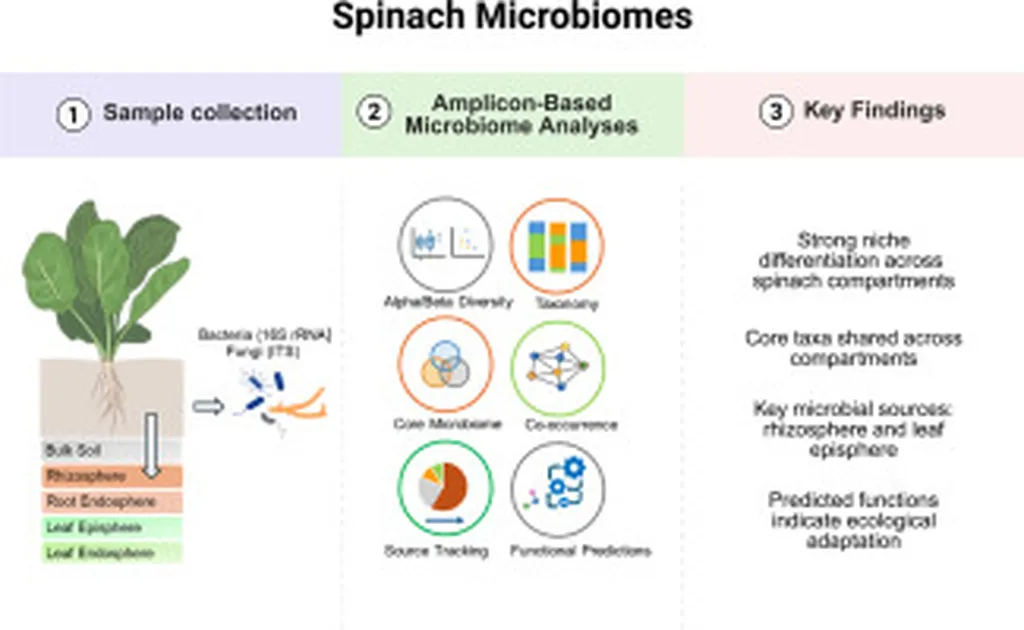
The study, led by Dhivya P. Thenappan at the Texas A&M AgriLife Research and Extension Center in Uvalde, Texas, examined microbial communities across five ecological niches: bulk soil, rhizosphere, leaf episphere, root endosphere, and leaf endosphere. Using 16S rRNA and ITS amplicon sequencing, the researchers identified significant cultivar-specific effects, particularly in the fungal community, and observed clear niche differentiation.
“Understanding the spatial organization and functional diversity of microbiomes across plant compartments is essential for unraveling the complex interactions between plants and their associated microbial consortia,” Thenappan explained. The research revealed that microbial diversity and community composition varied significantly across niches, with the highest diversity found in bulk soil, followed by the rhizosphere, leaf episphere, and finally the root and leaf endosphere.
The study identified a core microbiome comprising 10 bacterial and 6 fungal taxa, with Streptomyces, Bacillus, and Rubrobacter being the key bacterial genera, and Alternaria and Cladosporium the dominant fungi. SourceTracker2 analysis showed a limited contribution of bulk soil to the rhizosphere (∼25 %), while the rhizosphere and leaf episphere were the primary sources for endosphere communities. Fungal communities exhibited higher transfer rates (75–96 %) between niches compared to bacterial communities (19–93 %).
Co-occurrence network analysis revealed that the Traverse cultivar had a denser microbial network than Hammerhead, with key hubs such as Streptomycetaceae and Chaetomiaceae. Inferred functional potential suggested metabolic capabilities in microbial communities across spinach niches, including those of potential plant pathogens.
The findings lay the foundation for designing targeted strategies to mitigate pathogen risks and introduce beneficial microbial functions into crops. “This research provides a roadmap for future developments in the field, offering a deeper understanding of how microbial communities interact with spinach and how we can leverage this knowledge to improve agricultural practices,” Thenappan said.
The commercial impacts of this research are substantial. By identifying niche-specific microbial communities and their functional potential, farmers and agronomists can develop more effective strategies for crop management. This includes the potential to enhance plant health, increase yields, and reduce the reliance on chemical inputs, ultimately leading to more sustainable and profitable agricultural practices.
As the agricultural sector continues to face challenges such as climate change, soil degradation, and pest resistance, the insights gained from this study offer a promising avenue for innovation. By harnessing the power of microbial communities, the industry can move towards more resilient and productive farming systems.
The research, published in *Current Research in Microbial Sciences* and led by Dhivya P. Thenappan at the Texas A&M AgriLife Research and Extension Center, represents a significant step forward in our understanding of plant microbiomes and their potential applications in agriculture. As the field continues to evolve, the findings from this study will undoubtedly shape future developments and contribute to a more sustainable and productive agricultural sector.