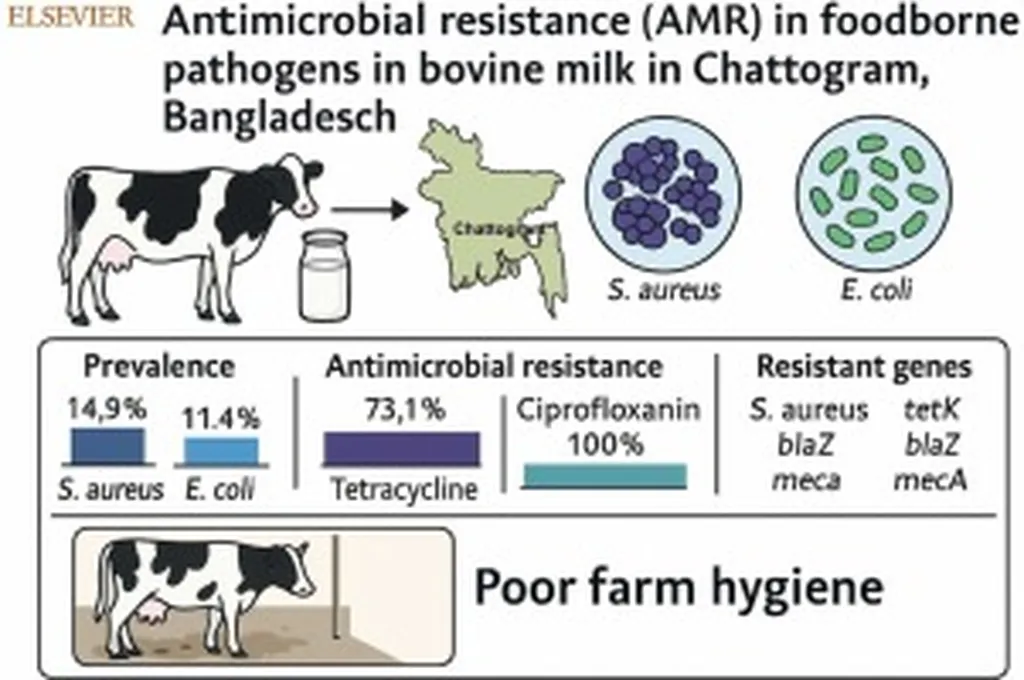
In the heart of Bangladesh’s dairy industry, a silent battle is raging—one that threatens both livestock and public health. Researchers have recently uncovered the genomic secrets of multidrug-resistant (MDR) Escherichia coli strains causing bovine mastitis, shedding light on their evolutionary pathways and potential zoonotic risks. This groundbreaking study, published in *BMC Microbiology*, could reshape our understanding of antimicrobial resistance and its impact on agriculture.
Bovine mastitis, a common and costly disease in dairy cattle, not only compromises animal welfare but also poses a significant economic burden. The emergence of MDR E. coli strains exacerbates this issue, as conventional treatments become less effective. Lead author Naim Siddique from the Molecular Biology and Bioinformatics Laboratory at Gazipur Agricultural University emphasizes the urgency of the situation: “Our findings highlight the critical need for One Health-based genomic surveillance to mitigate the transmission of MDR E. coli from dairy farms to humans and the environment.”
The study focuses on two MDR E. coli strains, MBBL4 and MBBL5, isolated from cases of bovine mastitis. Through comprehensive genomic analysis, researchers revealed that these strains exhibit extensive resistance to ten different antibiotics. Phylogenetic and average nucleotide identity (ANI) analyses showed that MBBL4 is closely related to strains associated with human bacteremia, while MBBL5 is linked to wildlife-associated strains. This divergence suggests that these bacteria have adapted to various hosts, increasing their potential for zoonotic transmission.
One of the most striking findings is the open pangenome structure of these strains, indicating high genetic diversity. MBBL4 harbors 21 unique genes, while MBBL5 possesses nine unique genes. This genetic variability underscores the adaptive potential of these bacteria, enabling them to thrive in diverse environments and resist multiple antibiotics.
The study also identified numerous antimicrobial resistance genes (ARGs) and virulence factor genes (VFGs) in both strains. These genes are predominantly associated with adherence and secretion systems, enhancing the bacteria’s ability to colonize and infect hosts. The presence of abundant mobile genetic elements (MGEs), such as plasmids, prophages, insertion sequence elements, and genomic islands, further highlights the role of horizontal gene transfer in driving resistance and virulence.
The commercial impact of these findings on the agriculture sector is profound. As Naim Siddique notes, “Understanding the genomic landscape of MDR E. coli in dairy cattle is crucial for developing targeted interventions and preventing the spread of resistance.” This research could lead to the development of new diagnostic tools, vaccines, and treatment strategies tailored to combat these resilient strains.
Moreover, the study underscores the importance of a One Health approach, which recognizes the interconnectedness of human, animal, and environmental health. By implementing genomic surveillance and control measures on dairy farms, the agriculture sector can mitigate the risk of MDR E. coli transmission, safeguarding both livestock and public health.
As the world grapples with the growing threat of antimicrobial resistance, this research provides valuable insights into the evolutionary dynamics of MDR E. coli. The findings not only highlight the urgent need for coordinated action but also pave the way for innovative solutions to protect the dairy industry and beyond. In the words of Naim Siddique, “This study is a stepping stone towards a more resilient and sustainable future for agriculture and public health.”