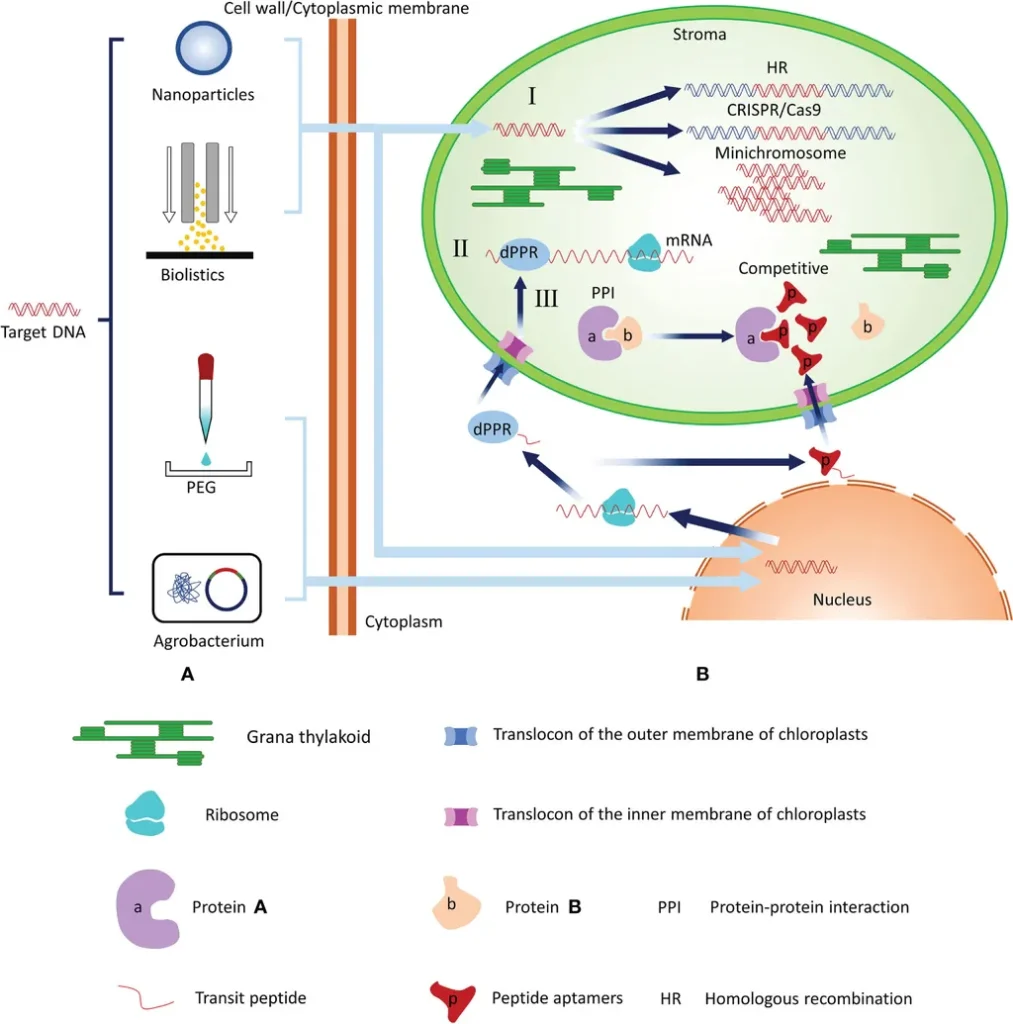
In a groundbreaking study published in *Frontiers in Plant Science*, researchers have harnessed the power of deep learning to decode the intricate architecture of chloroplast translational regulatory sequences. Led by Mohammad Ali Abbasi-Vineh from the Department of Cell & Molecular Biology at Shahid Beheshti University in Tehran, Iran, the research opens new avenues for synthetic biology and genetic engineering in both plants and algae.
The study introduces a hybrid deep learning model that combines convolutional neural networks (CNN), long short-term memory (LSTM) networks, Attention mechanisms, and Residual architectures. This sophisticated model was trained to classify and analyze two critical datasets: 5′ untranslated region sequences from plants and algae, and sequences with and without Shine-Dalgarno (SD) motifs. By focusing on 300-nucleotide leader sequences upstream of the start codon, the model achieved remarkable accuracy in predicting both the taxonomic origin of the sequences and the presence or absence of SD motifs.
One of the most intriguing findings was the identification of a small subset of plant and algal sequences that exhibited patterns typically associated with the other group. This discovery holds significant promise for the transfer of functional heterologous sequences between plants and algae, potentially revolutionizing genetic engineering efforts.
“Our model not only accurately classifies these sequences but also reveals subtle differences that could be exploited for cross-species applications,” said Abbasi-Vineh. “This opens up new possibilities for enhancing crop yields and improving algal biofuel production through targeted genetic modifications.”
The research highlights substantial differences in the plastid leader sequences between plants and algae, particularly within the first 30 base pairs upstream of the start codon. These findings suggest two promising strategies for algal plastome engineering: (1) using plant-derived leader sequences that mimic algal patterns, tailored to specific algal strains, and (2) constructing hybrid leader sequences by combining algae-specific upstream regions with plant-derived distal regions, incorporating SD motifs.
The commercial implications for the agriculture sector are vast. By optimizing chloroplast translation, researchers can enhance the efficiency of photosynthesis, improve crop resilience, and boost biofuel production from algae. These advancements could lead to more sustainable agricultural practices and contribute to food security in an era of climate change.
As the first deep learning model to analyze chloroplast translational regulatory sequences, this study provides a robust framework for future research. The findings offer valuable insights into the functional elements of chloroplast genomes, paving the way for innovative genetic engineering techniques that could transform both plant and algal biotechnology.
“This research is a significant step forward in our understanding of chloroplast translation,” noted Abbasi-Vineh. “It sets the stage for developing novel strategies that could have far-reaching impacts on agriculture and bioenergy production.”
With its potential to bridge the gap between plant and algal genetic engineering, this study not only advances scientific knowledge but also offers practical solutions for some of the most pressing challenges in the agriculture sector. As researchers continue to refine these deep learning models, the future of synthetic biology looks brighter than ever.