In the heart of agricultural innovation, a groundbreaking study published in the journal ‘Rice’ is set to revolutionize the way we approach rice cultivation. Led by Tilak Chandra from the Division of Agricultural Bioinformatics at ICAR-Indian Agricultural Statistics Research Institute, the research delves into the intricate world of non-coding DNA elements in rice, offering a novel approach to fine-tune agronomically advantageous traits.
Rice, a staple crop for nearly one-third of the global population, is under immense pressure to adapt to the changing climate and increasing food demands. Traditional breeding methods have long been the go-to strategy for enhancing crop yields, but they often fall short in addressing the complex challenges faced by modern agriculture. This is where the power of non-coding DNA elements comes into play.
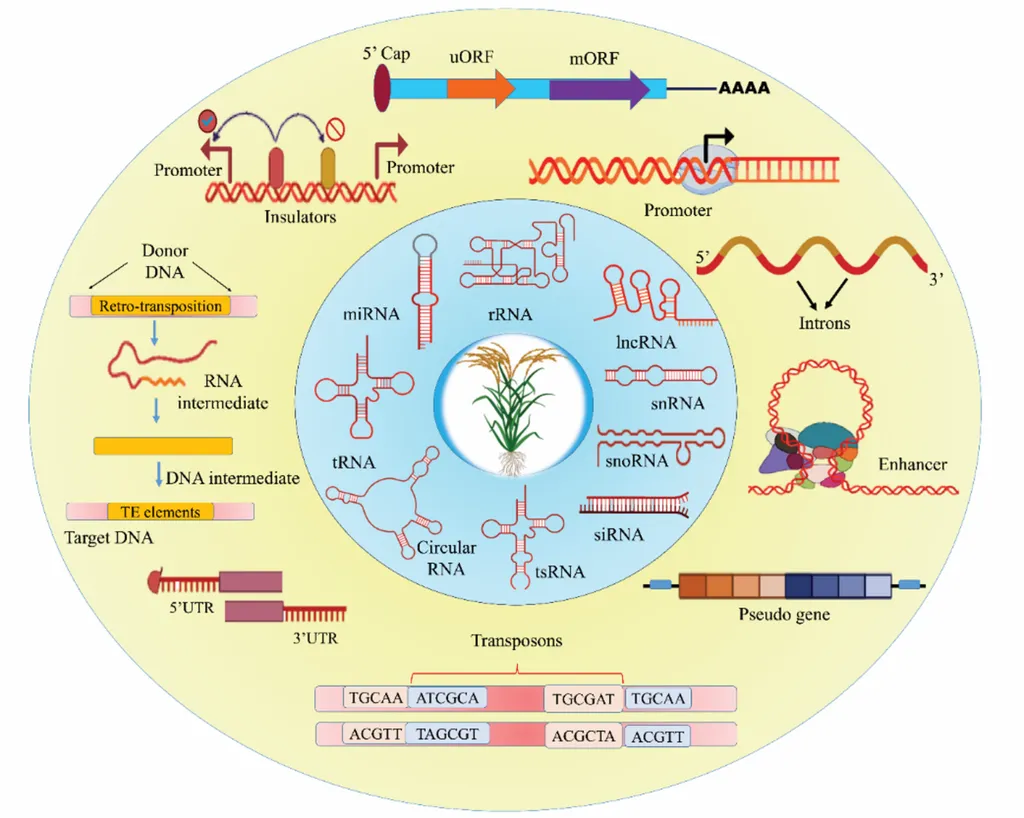
Non-coding DNA, once dismissed as “junk DNA,” has emerged as a critical player in regulating gene expression and orchestrating essential biological processes. Natural variations within these elements have been shown to contribute significantly to adaptive plasticity, domestication traits, and intraspecific diversification in rice. By targeting these non-coding elements, scientists can precisely modulate desirable agronomic traits, offering a more nuanced approach than traditional genetic modification.
“Non-coding elements are like the conductors of an orchestra,” explains Chandra. “They don’t produce the music themselves, but they regulate the performance of the genes, which are the musicians. By fine-tuning these conductors, we can enhance the overall performance of the crop.”
The study highlights the potential of engineered non-coding RNA elements for enhancing traits such as resilience to abiotic stresses, accelerated development, and resistance to biotic agents. This could lead to the development of rice varieties that are not only high-yielding but also more resilient to environmental stresses, ultimately contributing to global food and nutritional security.
The commercial implications of this research are vast. With the global rice market valued at over $250 billion, the demand for high-quality, high-yielding rice varieties is on the rise. The ability to precisely engineer non-coding elements offers a promising avenue for agricultural biotechnology companies to develop superior rice genotypes, catering to the evolving needs of farmers and consumers alike.
Moreover, the study underscores the importance of continuous technological innovations in the editing toolbox, such as CRISPR/Cas9, and breakthrough discoveries for non-coding elements influencing agronomical traits. These advances are expected to shape the future roadmap of non-coding element editing in rice, paving the way for a new era of agricultural biotechnology.
As we stand on the brink of a new agricultural revolution, the research led by Chandra offers a glimpse into the future of rice cultivation. By harnessing the power of non-coding DNA elements, we can unlock the full potential of this vital crop, ensuring sustainable food and nutritional security for generations to come. With the study published in ‘Rice’ and led by Tilak Chandra from the Division of Agricultural Bioinformatics at ICAR-Indian Agricultural Statistics Research Institute, the stage is set for a new chapter in agricultural innovation.