In the relentless battle against tuberculosis (TB), a formidable infectious disease that claims lives worldwide, researchers are turning to cutting-edge technology to stay ahead. A recent study published in the *Journal of Global Antimicrobial Resistance* has shed light on the promising potential of machine learning (ML) algorithms trained on genomic data to predict drug resistance in TB. This breakthrough could revolutionize the way we approach TB treatment, particularly in the agricultural sector where the disease poses significant challenges.
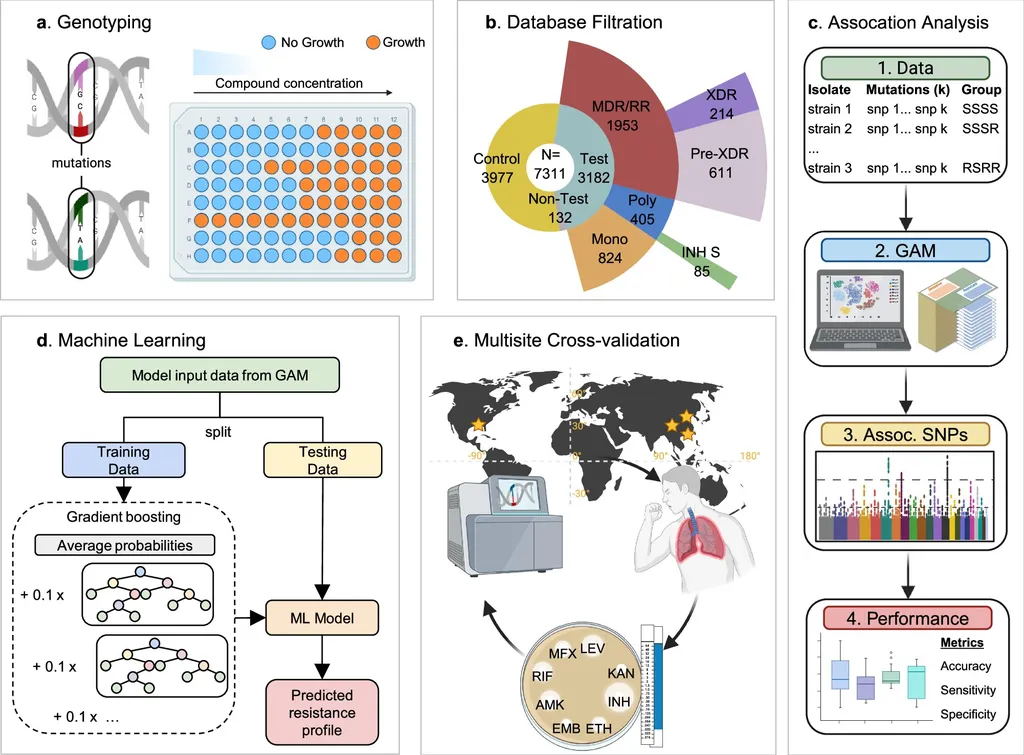
The study, led by Rohan Shrivastava from the Department of Translational Medicine at AIIMS Bhopal, Madhya Pradesh, India, conducted a systematic review and meta-analysis of seven original studies. These studies applied ML to whole-genome or targeted sequencing to predict resistance to first-line antitubercular drugs such as rifampicin (RIF), isoniazid (INH), ethambutol (EMB), and streptomycin (STM).
The findings are compelling. The pooled performance of ML algorithms was strongest for RIF and INH, with sensitivities of 0.90 and 0.88, and specificities of 0.95 and 0.93, respectively. “This indicates that ML models can accurately predict resistance to these drugs, which is crucial for guiding therapy and improving patient outcomes,” Shrivastava explained.
However, the performance varied for EMB and STM, with lower sensitivities despite reasonable AUCs. This variation underscores the complexity of TB drug resistance and the need for tailored approaches. “While the results are promising, there is still room for improvement, particularly in terms of sensitivity across different drugs and model types,” Shrivastava noted.
The study also highlighted the importance of standardized external validation and calibration before these models can be broadly deployed in clinical settings. This step is crucial to ensure the reliability and accuracy of the predictions, which could have significant implications for the agricultural sector.
In agriculture, TB poses a significant threat to livestock health and productivity. Rapid and accurate prediction of drug resistance can help farmers and veterinarians make informed decisions about treatment, thereby reducing the spread of the disease and improving animal welfare. Moreover, the use of ML and genomic data can streamline the diagnostic process, making it more efficient and cost-effective.
The commercial impact of this research could be substantial. By integrating ML algorithms into existing diagnostic tools, agricultural businesses can enhance their ability to manage TB outbreaks, reduce economic losses, and ensure the sustainability of their operations. Furthermore, the insights gained from this research can drive the development of new diagnostic tools and treatment strategies, opening up new avenues for innovation and investment in the agritech sector.
As we look to the future, the potential of ML in predicting drug resistance is immense. This research not only highlights the importance of integrating advanced technologies into healthcare and agriculture but also paves the way for further exploration and development in this field. By continuing to refine and validate these models, we can unlock new possibilities for combating TB and other infectious diseases, ultimately improving the lives of both humans and animals.