In a groundbreaking study published in *Nature Communications*, researchers have unraveled the genetic mystery behind the mid-oleic fatty acid phenotype in peanuts, a trait highly valued for its health benefits and commercial potential. The research, led by Ethan Thompson from the Department of Plant Pathology at the University of Georgia, challenges the traditional two-gene model and introduces a novel third gene, AhFAD2C, which plays a crucial role in determining the mid-oleic fatty acid content in peanuts.
For years, scientists have relied on a two-gene model involving AhFAD2A and AhFAD2B to explain the mid-oleic fatty acid phenotype. However, accumulating evidence suggested that this model was incomplete. “The traditional two-gene model couldn’t fully account for the observed phenotypic variation,” Thompson explains. “This discrepancy hinted at the existence of additional genetic factors.”
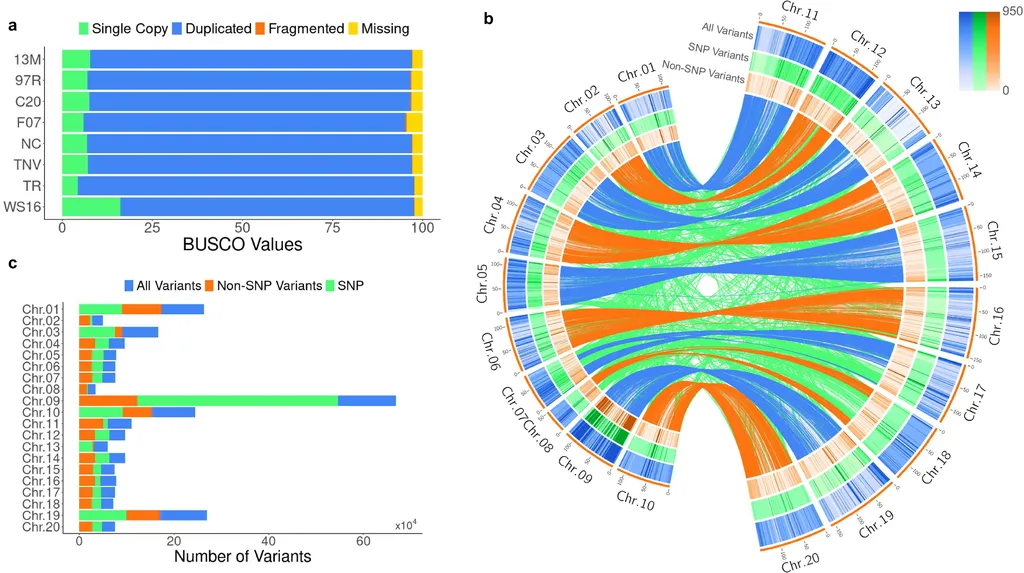
To address this puzzle, Thompson and his team constructed a population-specific pangenome using the eight founder genomes of the PeanutMAGIC population. This graph-based pangenome serves as a comprehensive reference, capturing all segregating haplotypes within the population. By conducting whole-genome sequencing on the MAGIC Core, a subset of 310 recombinant inbred lines (RILs), the researchers were able to trace recombination events and perform detailed genomic analyses.
The study identified a unique third gene, AhFAD2C, located near AhFAD2B. When recombination occurs, AhFAD2C segregates from AhFAD2B, revealing a previously unknown genetic mechanism. “The identification of AhFAD2C fills a critical gap in our understanding of the genetic architecture underlying the mid-oleic fatty acid phenotype,” Thompson states.
The implications of this research are significant for the agriculture sector. Peanuts with mid-oleic fatty acid content are highly sought after for their health benefits, including improved heart health and longer shelf life. Understanding the genetic basis of this trait allows breeders to develop peanut varieties with enhanced oil quality more efficiently. “This discovery opens up new avenues for breeding programs aimed at improving peanut oil quality,” Thompson notes.
Moreover, the study highlights the limitations of relying on a single-reference genome. Traditional approaches can lead to false associations and marker discovery, whereas a population-specific pangenome provides a more reliable framework for genomic studies. This insight could revolutionize how genetic research is conducted in other crops, leading to more accurate and comprehensive genetic analyses.
The research not only solves a long-standing mystery in peanut genetics but also demonstrates the power of population-specific pangenomes in uncovering hidden genetic mechanisms. As Thompson and his team continue to explore the genetic diversity within peanut populations, their work is poised to shape the future of peanut breeding and the broader field of agricultural genomics.